Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon β
- PMID: 21862579
- PMCID: PMC3195586
- DOI: 10.1074/jbc.M111.267567
Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon β
Abstract
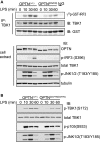
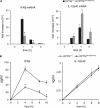
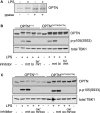
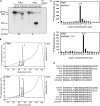
TANK-binding kinase (TBK1) is essential for transcription of the interferon (IFN) β gene in response to lipopolysaccharide (LPS) and double-stranded RNA, but the molecular mechanisms that underlie the activation of TBK1 are incompletely understood. Previously, we identified the NF-κB essential modulator (NEMO)-related polyubiquitin-binding protein, optineurin (OPTN), as a novel binding partner of TBK1. To determine whether the ubiquitin-binding function of OPTN is involved in regulating TBK1 and IFNβ production, we generated a mouse in which wild-type optineurin was replaced by the polyubiquitin binding-defective mutant, OPTN(D477N/D477N). In this study, we found that LPS or poly(I:C)-induced TBK1 activity was significantly reduced in bone marrow-derived macrophage (BMDM) from OPTN(D477N/D477N) mice. Consistent with this, the phosphorylation of IFN regulatory factor 3 (IRF3) and the production of IFNβ mRNA and secretion were reduced. Stimulation of BMDMs with LPS triggered the phosphorylation of OPTN, which was reversed by phosphatase treatment and prevented by pharmacological inhibition of both the canonical IκB kinases (IKKα/β) and the IKK-related kinases (TBK1/IKKε). In contrast, LPS-stimulated phosphorylation of OPTN(D477N) was markedly reduced in BMDMs from OPTN(D477N/D477N) mice, and inhibition of the canonical IKKs alone prevented phosphorylation, providing further evidence that ubiquitin binding to OPTN contributes to LPS-induced TBK1 activation. TBK1 and IKKβ phosphorylated OPTN preferentially at Ser-177 and Ser-513, respectively, in vitro. In conclusion, our results suggest that OPTN binds to polyubiquitylated species formed in response to LPS and poly(I:C), enhancing the activation of TBK1 that is required for optimal phosphorylation of IRF3 and production of IFNβ.
Figures




References
-
- Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) Nature 412, 346–351 - PubMed
-
- Haas T. L., Emmerich C. H., Gerlach B., Schmukle A. C., Cordier S. M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T., Koschny R., Komander D., Silke J., Walczak H. (2009) Mol. Cell 36, 831–844 - PubMed
-
- Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., Iwai K. (2009) Nat. Cell Biol. 11, 123–132 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

