A molecular chaperone mediates a two-protein enzyme complex and glycosylation of serine-rich streptococcal adhesins
- PMID: 21862581
- PMCID: PMC3186434
- DOI: 10.1074/jbc.M111.239350
A molecular chaperone mediates a two-protein enzyme complex and glycosylation of serine-rich streptococcal adhesins
Abstract
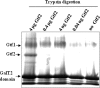
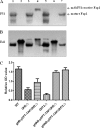
Serine-rich repeat glycoproteins identified from streptococci and staphylococci are important for bacterial adhesion and biofilm formation. Two putative glycosyltransferases, Gtf1 and Gtf2, from Streptococcus parasanguinis form a two-protein enzyme complex that is required for glycosylation of a serine-rich repeat adhesin, Fap1. Gtf1 is a glycosyltransferase; however, the function of Gtf2 is unknown. Here, we demonstrate that Gtf2 enhances the enzymatic activity of Gtf1 by its chaperone-like property. Gtf2 interacted with Gtf1, mediated the subcellular localization of Gtf1, and stabilized Gtf1. Deletion of invariable amino acid residues in a conserved domain of unknown function (DUF1975) at the N terminus of Gtf2 had a greater impact on Fap1 glycosylation than deletion of the C-terminal non-DUF1975 residues. The DUF1975 deletions concurrently reduced the interaction between Gtf1 and Gtf2, altered the subcellular localization of Gtf1, and destabilized Gtf1, suggesting that DUF1975 is crucial for the chaperone activity of Gtf2. Homologous GtfA and GtfB from Streptococcus agalactiae rescued the glycosylation defect in the gtf1gtf2 mutant; like Gtf2, GtfB also possesses chaperone-like activity. Taken together, our studies suggest that Gtf2 and its homologs possess the conserved molecular chaperone activity that mediates protein glycosylation of bacterial adhesins.
Figures







Similar articles
-
Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin.J Bacteriol. 2008 Feb;190(4):1256-66. doi: 10.1128/JB.01078-07. Epub 2007 Dec 14. J Bacteriol. 2008. PMID: 18083807 Free PMC article.
-
Engineering and Dissecting the Glycosylation Pathway of a Streptococcal Serine-rich Repeat Adhesin.J Biol Chem. 2016 Dec 30;291(53):27354-27363. doi: 10.1074/jbc.M116.752998. J Biol Chem. 2016. PMID: 28039332 Free PMC article.
-
A novel glucosyltransferase is required for glycosylation of a serine-rich adhesin and biofilm formation by Streptococcus parasanguinis.J Biol Chem. 2010 Apr 16;285(16):12140-8. doi: 10.1074/jbc.M109.066928. Epub 2010 Feb 17. J Biol Chem. 2010. PMID: 20164186 Free PMC article.
-
Glycosylation and biogenesis of a family of serine-rich bacterial adhesins.Microbiology (Reading). 2009 Feb;155(Pt 2):317-327. doi: 10.1099/mic.0.025221-0. Microbiology (Reading). 2009. PMID: 19202081 Review.
-
Glycosyltransferase-mediated Sweet Modification in Oral Streptococci.J Dent Res. 2015 May;94(5):659-65. doi: 10.1177/0022034515574865. Epub 2015 Mar 9. J Dent Res. 2015. PMID: 25755271 Free PMC article. Review.
Cited by
-
New Helical Binding Domain Mediates a Glycosyltransferase Activity of a Bifunctional Protein.J Biol Chem. 2016 Oct 14;291(42):22106-22117. doi: 10.1074/jbc.M116.731695. Epub 2016 Aug 17. J Biol Chem. 2016. PMID: 27539847 Free PMC article.
-
Both GtfA and GtfB are required for SraP glycosylation in Staphylococcus aureus.Curr Microbiol. 2014 Aug;69(2):121-6. doi: 10.1007/s00284-014-0563-2. Curr Microbiol. 2014. PMID: 24658735
-
The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold.Nat Commun. 2014 Jul 15;5:4339. doi: 10.1038/ncomms5339. Nat Commun. 2014. PMID: 25023666 Free PMC article.
-
Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.Acta Crystallogr D Struct Biol. 2021 Nov 1;77(Pt 11):1460-1474. doi: 10.1107/S2059798321010068. Epub 2021 Oct 20. Acta Crystallogr D Struct Biol. 2021. PMID: 34726173 Free PMC article.
-
Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein.Proc Natl Acad Sci U S A. 2016 Mar 1;113(9):E1190-9. doi: 10.1073/pnas.1600494113. Epub 2016 Feb 16. Proc Natl Acad Sci U S A. 2016. PMID: 26884191 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

