InvA protein is a Nudix hydrolase required for infection by pathogenic Leptospira in cell lines and animals
- PMID: 21862592
- PMCID: PMC3196074
- DOI: 10.1074/jbc.M111.219931
InvA protein is a Nudix hydrolase required for infection by pathogenic Leptospira in cell lines and animals
Retraction in
-
Retraction. InvA protein is a Nudix hydrolase required for infection by pathogenic Leptospira in cell lines and animals.J Biol Chem. 2012 Mar 16;287(12):9327. doi: 10.1074/jbc.a111.219931. J Biol Chem. 2012. PMID: 22532973 Free PMC article. No abstract available.
Abstract
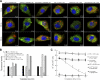
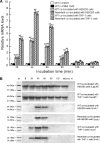
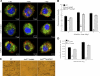
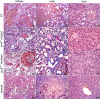
Leptospirosis caused by pathogenic species of the genus Leptospira is a re-emerging zoonotic disease, which affects a wide variety of host species and is transmitted by contaminated water. The genomes of several pathogenic Leptospira species contain a gene named invA, which contains a Nudix domain. However, the function of this gene has never been characterized. Here, we demonstrated that the invA gene was highly conserved in protein sequence and present in all tested pathogenic Leptospira species. The recombinant InvA protein of pathogenic L. interrogans strain Lai hydrolyzed several specific dinucleoside oligophosphate substrates, reflecting the enzymatic activity of Nudix in Leptospira species. Pathogenic leptospires did not express this protein in media but temporarily expressed it at early stages (within 60 min) of infection of macrophages and nephric epithelial cells. Comparing with the wild type, the invA-deficient mutant displayed much lower infectivity and a significantly reduced survival rate in macrophages and nephric epithelial cells. Moreover, the invA-deficient leptospires presented an attenuated virulence in hamsters, caused mild histopathological damage, and were transmitted in lower numbers in the urine, compared with the wild-type strain. The invA revertant, made by complementing the invA-deficient mutant with the invA gene, reacquired virulence similar to the wild type in vitro and in vivo. The LD(50) in hamsters was 1000-fold higher for the invA-deficient mutant than for the invA revertant and wild type. These results demonstrate that the InvA protein is a Nudix hydrolase, and the invA gene is essential for virulence in pathogenic Leptospira species.
Figures

References
-
- Bharti A. R., Nally J. E., Ricaldi J. N., Matthias M. A., Diaz M. M., Lovett M. A., Levett P. N., Gilman R. H., Willig M. R., Gotuzzo E., Vinetz J. M. (2003) Lancet Infect. Dis. 3, 757–771 - PubMed
-
- Vijayachari P., Sugunan A. P., Shriram A. N. (2008) J. Biosci. 33, 557–569 - PubMed
-
- Djadid N. D., Ganji Z. F., Gouya M. M., Rezvani M., Zakeri S. (2009) Diagn. Microbiol. Infect. Dis. 63, 251–256 - PubMed
-
- Chidambaram N., Ramanathan M., Anandi V., Sasikala S., Innocent D. J., Sarayu L. (2007) J. Commun. Dis. 39, 105–108 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

