Fluorescence image screening for chemical compounds modifying cholesterol metabolism and distribution
- PMID: 21862703
- PMCID: PMC3196239
- DOI: 10.1194/jlr.D018184
Fluorescence image screening for chemical compounds modifying cholesterol metabolism and distribution
Abstract
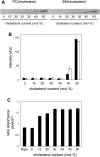
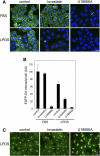
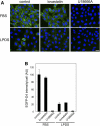
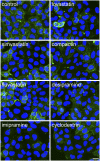
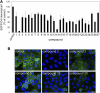
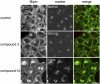
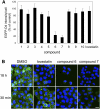
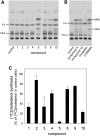
An automated fluorescence microscopy assay using a nontoxic cholesterol binding protein, toxin domain 4, (D4), was developed in order to identify chemical compounds modifying intracellular cholesterol metabolism and distribution. Using this method, we screened a library of 1,056 compounds and identified 35 compounds that decreased D4 binding to the cell surface. Among them, 8 compounds were already reported to alter the biosynthesis or the intracellular distribution of cholesterol. The remaining 27 hit compounds were further analyzed biochemically and histochemically. Cell staining with another fluorescent cholesterol probe, filipin, revealed that 17 compounds accumulated cholesterol in the late endosomes. Five compounds decreased cholesterol biosynthesis, and two compounds inhibited the binding of D4 to the membrane. This visual screening method, based on the cholesterol-specific probe D4 in combination with biochemical analyses, is a cell-based, sensitive technique for identifying new chemical compounds and modifying cholesterol distribution and metabolism. Furthermore, it is suitable for high-throughput analysis for drug discovery.
Figures

References
-
- Simons K., Vaz W. L. 2004. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33: 269–295 - PubMed
-
- Epand R. M., Thomas A., Brasseur R., Epand R. F. 2010. Cholesterol interaction with proteins that partition into membrane domains: an overview. Subcell. Biochem. 51: 253–278 - PubMed
-
- Simons K., Toomre D. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1: 31–39 - PubMed
-
- Goldstein J. L., DeBose-Boyd R. A., Brown M. S. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46 - PubMed
-
- Ikonen E. 2008. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 9: 125–138 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

