RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis
- PMID: 21863050
- PMCID: PMC3542975
- DOI: 10.1038/nrc3120
RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis
Abstract
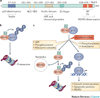
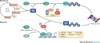
The ubiquitin-proteasome system has numerous crucial roles in physiology and pathophysiology. Fundamental to the specificity of this system are ubiquitin-protein ligases (E3s). Of these, the majority are RING finger and RING finger-related E3s. Many RING finger E3s have roles in processes that are central to the maintenance of genomic integrity and cellular homeostasis, such as the anaphase promoting complex/cyclosome (APC/C), the SKP1-cullin 1-F-box protein (SCF) E3s, MDM2, BRCA1, Fanconi anaemia proteins, CBL proteins, von Hippel-Lindau tumour suppressor (VHL) and SIAH proteins. As a result, many RING finger E3s are implicated in either the suppression or the progression of cancer. This Review summarizes current knowledge in this area.
Figures

References
-
-
Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)- dependent ubiquitination. Proc. Natl. Acad. Sci. U S A. 1999;96:11364–11369. This study determined that RING fingers are E2-binding ubiquitin ligase domains and that RING finger-containing proteins including SIAH and BRCA1 have ubiquitin ligase activity.
-
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6:369–381. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

