HIV-1 protease inhibitor mutations affect the development of HIV-1 resistance to the maturation inhibitor bevirimat
- PMID: 21864346
- PMCID: PMC3184055
- DOI: 10.1186/1742-4690-8-70
HIV-1 protease inhibitor mutations affect the development of HIV-1 resistance to the maturation inhibitor bevirimat
Abstract
Background: Maturation inhibitors are an experimental class of antiretrovirals that inhibit Human Immunodeficiency Virus (HIV) particle maturation, the structural rearrangement required to form infectious virus particles. This rearrangement is triggered by the ordered cleavage of the precursor Gag polyproteins into their functional counterparts by the viral enzyme protease. In contrast to protease inhibitors, maturation inhibitors impede particle maturation by targeting the substrate of protease (Gag) instead of the protease enzyme itself. Direct cross-resistance between protease and maturation inhibitors may seem unlikely, but the co-evolution of protease and its substrate, Gag, during protease inhibitor therapy, could potentially affect future maturation inhibitor therapy. Previous studies showed that there might also be an effect of protease inhibitor resistance mutations on the development of maturation inhibitor resistance, but the exact mechanism remains unclear. We used wild-type and protease inhibitor resistant viruses to determine the impact of protease inhibitor resistance mutations on the development of maturation inhibitor resistance.
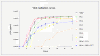
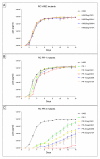
Results: Our resistance selection studies demonstrated that the resistance profiles for the maturation inhibitor bevirimat are more diverse for viruses with a mutated protease compared to viruses with a wild-type protease. Viral replication did not appear to be a major factor during emergence of bevirimat resistance. In all in vitro selections, one of four mutations was selected: Gag V362I, A364V, S368N or V370A. The impact of these mutations on maturation inhibitor resistance and viral replication was analyzed in different protease backgrounds. The data suggest that the protease background affects development of HIV-1 resistance to bevirimat and the replication profiles of bevirimat-selected HIV-1. The protease-dependent bevirimat resistance and replication levels can be explained by differences in CA/p2 cleavage processing by the different proteases.
Conclusions: These findings highlight the complicated interactions between the viral protease and its substrate. By providing a better understanding of these interactions, we aim to help guide the development of second generation maturation inhibitors.
Figures
References
-
- Nijhuis M, van Maarseveen NM, Lastere S, Schipper P, Coakley E, Glass B, Rovenska M, de Jong D, Chappey C, Goedegebuure IW, Heilek-Snyder G, Dulude D, Cammack N, Brakier-Gingras L, Konvalinka J, Parkin N, Krausslich HG, Brun-Vezinet F, Boucher CA. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007;4:e36. doi: 10.1371/journal.pmed.0040036. - DOI - PMC - PubMed
-
- Verheyen J, Litau E, Sing T, Daumer M, Balduin M, Oette M, Fatkenheuer G, Rockstroh JK, Schuldenzucker U, Hoffmann D, Pfister H, Kaiser R. Compensatory mutations at the HIV cleavage sites p7/p1 and p1/p6-gag in therapy-naive and therapy-experienced patients. Antivir Ther. 2006;11:879–887. - PubMed
-
- Dam E, Quercia R, Glass B, Descamps D, Launay O, Duval X, Krausslich HG, Hance AJ, Clavel F. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 2009;5:e1000345. doi: 10.1371/journal.ppat.1000345. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

