Whole-genome phylogenies of the family Bacillaceae and expansion of the sigma factor gene family in the Bacillus cereus species-group
- PMID: 21864360
- PMCID: PMC3171730
- DOI: 10.1186/1471-2164-12-430
Whole-genome phylogenies of the family Bacillaceae and expansion of the sigma factor gene family in the Bacillus cereus species-group
Abstract
Background: The Bacillus cereus sensu lato group consists of six species (B. anthracis, B. cereus, B. mycoides, B. pseudomycoides, B. thuringiensis, and B. weihenstephanensis). While classical microbial taxonomy proposed these organisms as distinct species, newer molecular phylogenies and comparative genome sequencing suggests that these organisms should be classified as a single species (thus, we will refer to these organisms collectively as the Bc species-group). How do we account for the underlying similarity of these phenotypically diverse microbes? It has been established for some time that the most rapidly evolving and evolutionarily flexible portions of the bacterial genome are regulatory sequences and transcriptional networks. Other studies have suggested that the sigma factor gene family of these organisms has diverged and expanded significantly relative to their ancestors; sigma factors are those portions of the bacterial transcriptional apparatus that control RNA polymerase recognition for promoter selection. Thus, examining sigma factor divergence in these organisms would concurrently examine both regulatory sequences and transcriptional networks important for divergence. We began this examination by comparison to the sigma factor gene set of B. subtilis.
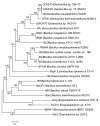
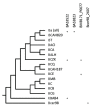
Results: Phylogenetic analysis of the Bc species-group utilizing 157 single-copy genes of the family Bacillaceae suggests that several taxonomic revisions of the genus Bacillus should be considered. Within the Bc species-group there is little indication that the currently recognized species form related sub-groupings, suggesting that they are members of the same species. The sigma factor gene family encoded by the Bc species-group appears to be the result of a dynamic gene-duplication and gene-loss process that in previous analyses underestimated the true heterogeneity of the sigma factor content in the Bc species-group.
Conclusions: Expansion of the sigma factor gene family appears to have preferentially occurred within the extracytoplasmic function (ECF) sigma factor genes, while the primary alternative (PA) sigma factor genes are, in general, highly conserved with those found in B. subtilis. Divergence of the sigma-controlled transcriptional regulons among various members of the Bc species-group likely has a major role in explaining the diversity of phenotypic characteristics seen in members of the Bc species-group.
Figures


References
-
- Rooney AP, Price NP, Ehrhardt C, Swezey JL, Bannan JD. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol. 2009;59(Pt 10):2429–2436. - PubMed
-
- Soufiane B, Cote JC. Bacillus thuringiensis Serovars bolivia, vazensis and navarrensis Meet the Description of Bacillus weihenstephanensis. Curr Microbiol. 2009. - PubMed
-
- Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolsto AB. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis--one species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66(6):2627–2630. doi: 10.1128/AEM.66.6.2627-2630.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

