Design and validation of consensus-degenerate hybrid oligonucleotide primers for broad and sensitive detection of corona- and toroviruses
- PMID: 21864579
- PMCID: PMC7112876
- DOI: 10.1016/j.jviromet.2011.08.005
Design and validation of consensus-degenerate hybrid oligonucleotide primers for broad and sensitive detection of corona- and toroviruses
Abstract
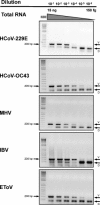
The ssRNA+ family Coronaviridae includes two subfamilies prototyped by coronaviruses and toroviruses that cause respiratory and enteric infections. To facilitate the identification of new distantly related members of the family Coronaviridae, we have developed a molecular assay with broad specificity. The consensus-degenerated hybrid oligonucleotide primer (CODEHOP) strategy was modified to design primers targeting the most conserved motifs in the RNA-dependent RNA polymerase locus. They were evaluated initially on RNA templates from virus-infected cells using a two-step RT-PCR protocol that was further advanced to a one-step assay. The sensitivity of the assay ranged from 10(2) to 10(6) and from 10(5) to 10(9) RNA copy numbers for individual corona-/torovirus templates when tested, respectively, with and without an excess of RNA from human cells. This primer set compared to that designed according to the original CODEHOP rules showed 10-10(3) folds greater sensitivity for 5 of the 6 evaluated corona-/torovirus templates. It detected 57% (32 of 56) of the respiratory specimens positive for 4 human coronaviruses, as well as stool specimens positive for a bovine torovirus. The high sensitivity and broad virus range of this assay makes it suitable for screening biological specimens in search for new viruses of the family Coronaviridae.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures





References
-
- Baines J.E., McGovern R.M., Persing D., Gostout B.S. Consensus-degenerate hybrid oligonucleotide primers (CODEHOP) for the detection of novel papillomaviruses and their application to esophageal and tonsillar carcinomas. J. Virol. Methods. 2005;123:81–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

