Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase
- PMID: 21865075
- PMCID: PMC3189298
- DOI: 10.1016/j.cbpa.2011.08.003
Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase
Abstract
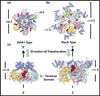
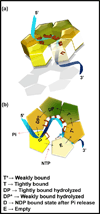
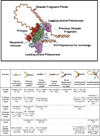
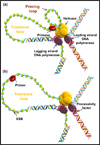
Helicases are molecular motor proteins that couple NTP hydrolysis to directional movement along nucleic acids. A class of helicases characterized by their ring-shaped hexameric structures translocate processively and unidirectionally along single-stranded (ss) DNA to separate the strands of double-stranded (ds) DNA, aiding both in the initiation and fork progression during DNA replication. These replicative ring-shaped helicases are found from virus to human. We review recent biochemical and structural studies that have expanded our understanding on how hexameric helicases use the NTPase reaction to translocate on ssDNA, unwind dsDNA, and how their physical and functional interactions with the DNA polymerase and primase enzymes coordinate replication of the two strands of dsDNA.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
References
-
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. - PubMed
-
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. - PubMed
-
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. - PubMed
-
- Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

