Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE
- PMID: 21865298
- PMCID: PMC3196886
- DOI: 10.1093/hmg/ddr363
Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE
Abstract
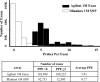
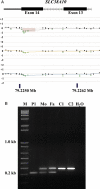
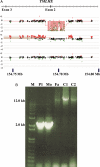
Autism is a neurodevelopmental disorder with increasing evidence of heterogeneous genetic etiology including de novo and inherited copy number variants (CNVs). We performed array comparative genomic hybridization using a custom Agilent 1 M oligonucleotide array intended to cover 197 332 unique exons in RefSeq genes; 98% were covered by at least one probe and 95% were covered by three or more probes with the focus on detecting relatively small CNVs that would implicate a single protein-coding gene. The study group included 99 trios from the Simons Simplex Collection. The analysis identified and validated 55 potentially pathogenic CNVs, categorized as de novo autosomal heterozygous, inherited homozygous autosomal, complex autosomal and hemizygous deletions on the X chromosome of probands. Twenty percent (11 of 55) of these CNV calls were rare when compared with the Database of Genomic Variants. Thirty-six percent (20 of 55) of the CNVs were also detected in the same samples in an independent analysis using the 1 M Illumina single-nucleotide polymorphism array. Findings of note included a common and sometimes homozygous 61 bp exonic deletion in SLC38A10, three CNVs found in lymphoblast-derived DNA but not present in whole-blood derived DNA and, most importantly, in a male proband, an exonic deletion of the TMLHE (trimethyllysine hydroxylase epsilon) that encodes the first enzyme in the biosynthesis of carnitine. Data for CNVs present in lymphoblasts but absent in fresh blood DNA suggest that these represent clonal outgrowth of individual B cells with pre-existing somatic mutations rather than artifacts arising in cell culture. GEO accession number GSE23765 (http://www.ncbi.nlm.nih.gov/geo/, date last accessed on 30 August 2011). Genboree accession: http://genboree.org/java-bin/gbrowser.jsp?refSeqId=1868&entryPointId=chr17&from=53496072&to=53694382&isPublic=yes, date last accessed on 30 August 2011.
Figures

References
-
- Jacquemont M.-L., Sanlaville D., Redon R., Raoul O., Cormier-Daire V., Lyonnet S., Amiel J., Le M.M., Heron D., De Blois M.-C., et al. Array-based comparative genomic hybridization identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J. Med. Genet. 2006;43:843–849. - PMC - PubMed
-
- Miles J.H., Takahashi T.N., Hong J., Munden N., Flournoy N., Braddock S.R., Martin R.A., Bocian M.E., Spence M.A., Hillman R.E., Farmer J.E. Development and validation of a measure of dysmorphology: useful for autism subgroup classification. Am. J. Med. Genet. A. 2008;146A:1101–1116. - PubMed
-
- Scott F.J., Baron-Cohen S., Bolton P., Brayne C. Brief report: prevalence of autism spectrum conditions in children aged 5–11 years in Cambridgeshire, UK. Autism. 2002;6:231–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

