Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL)
- PMID: 21865341
- PMCID: PMC3342856
- DOI: 10.1182/blood-2011-06-358382
Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL)
Abstract
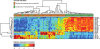
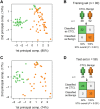
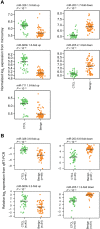
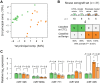
Cutaneous T-cell lymphomas (CTCLs) are the most frequent primary skin lymphomas. Nevertheless, diagnosis of early disease has proven difficult because of a clinical and histologic resemblance to benign inflammatory skin diseases. To address whether microRNA (miRNA) profiling can discriminate CTCL from benign inflammation, we studied miRNA expression levels in 198 patients with CTCL, peripheral T-cell lymphoma (PTL), and benign skin diseases (psoriasis and dermatitis). Using microarrays, we show that the most induced (miR-326, miR-663b, and miR-711) and repressed (miR-203 and miR-205) miRNAs distinguish CTCL from benign skin diseases with > 90% accuracy in a training set of 90 samples and a test set of 58 blinded samples. These miRNAs also distinguish malignant and benign lesions in an independent set of 50 patients with PTL and skin inflammation and in experimental human xenograft mouse models of psoriasis and CTCL. Quantitative (q)RT-PCR analysis of 103 patients with CTCL and benign skin disorders validates differential expression of 4 of the 5 miRNAs and confirms previous reports on miR-155 in CTCL. A qRT-PCR-based classifier consisting of miR-155, miR-203, and miR-205 distinguishes CTCL from benign disorders with high specificity and sensitivity, and with a classification accuracy of 95%, indicating that miRNAs have a high diagnostic potential in CTCL.
Figures


Comment in
-
Genetics: New miRNA classifier distinguishes CTCL from benign skin disorders.Nat Rev Clin Oncol. 2011 Sep 20;8(11):629. doi: 10.1038/nrclinonc.2011.144. Nat Rev Clin Oncol. 2011. PMID: 21931355 No abstract available.
-
Early CTCL diagnosis, a (miR)age no more?Blood. 2011 Nov 24;118(22):5717-8. doi: 10.1182/blood-2011-09-379107. Blood. 2011. PMID: 22123905 No abstract available.
References
-
- Trautinger F, Knobler R, Willemze R, et al. EORTC consensus recommendations for the treatment of mycosis fungoides/Sézary syndrome. Eur J Cancer. 2006;42(8):1014–1030. - PubMed
-
- Gjerdrum LM, Woetmann A, Odum N, et al. FOXP3+ regulatory T cells in cutaneous T-cell lymphomas association with disease stage and survival. Leukemia. 2007;21(12):2512–2518. - PubMed
-
- Rosen ST, Querfeld C. Primary cutaneous T-cell lymphomas. Hematol Am Soc Hematol Educ Program. 2006;323:330. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

