Ligand activation leads to regulated intramembrane proteolysis of fibroblast growth factor receptor 3
- PMID: 21865593
- PMCID: PMC3192865
- DOI: 10.1091/mbc.E11-01-0080
Ligand activation leads to regulated intramembrane proteolysis of fibroblast growth factor receptor 3
Abstract
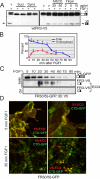
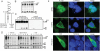
Fibroblast growth factor receptor 3 (FGFR3) is a major negative regulator of bone growth that inhibits the proliferation and differentiation of growth plate chondrocytes. Activating mutations of its c isoform cause dwarfism in humans; somatic mutations can drive oncogenic transformation in multiple myeloma and bladder cancer. How these distinct activities arise is not clear. FGFR3 was previously shown to undergo proteolytic cleavage in the bovine rib growth plate, but this was not explored further. Here, we show that FGF1 induces regulated intramembrane proteolysis (RIP) of FGFR3. The ectodomain is proteolytically cleaved (S1) in response to ligand-induced receptor activation, but unlike most RIP target proteins, it requires endocytosis and does not involve a metalloproteinase. S1 cleavage generates a C-terminal domain fragment that initially remains anchored in the membrane, is phosphorylated, and is spatially distinct from the intact receptor. Ectodomain cleavage is followed by intramembrane cleavage (S2) to generate a soluble intracellular domain that is released into the cytosol and can translocate to the nucleus. We identify the S1 cleavage site and show that γ-secretase mediates the S2 cleavage event. In this way we demonstrate a mechanism for the nuclear localization of FGFR3 in response to ligand activation, which may occur in both development and disease.
Figures
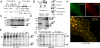



Similar articles
-
Fibroblast growth factors 1, 2, 17, and 19 are the predominant FGF ligands expressed in human fetal growth plate cartilage.Pediatr Res. 2007 Mar;61(3):267-72. doi: 10.1203/pdr.0b013e318030d157. Pediatr Res. 2007. PMID: 17314681
-
The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments.J Biol Chem. 2003 Sep 5;278(36):34427-37. doi: 10.1074/jbc.M302659200. Epub 2003 Jun 25. J Biol Chem. 2003. PMID: 12826675
-
Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and γ-secretase-dependent intramembranous cleavage.J Biol Chem. 2013 Nov 15;288(46):33027-36. doi: 10.1074/jbc.M113.517540. Epub 2013 Sep 27. J Biol Chem. 2013. PMID: 24078628 Free PMC article.
-
Achondroplasia: Development, pathogenesis, and therapy.Dev Dyn. 2017 Apr;246(4):291-309. doi: 10.1002/dvdy.24479. Epub 2017 Mar 2. Dev Dyn. 2017. PMID: 27987249 Free PMC article. Review.
-
Got RIP? Presenilin-dependent intramembrane proteolysis in growth factor receptor signaling.Cytokine Growth Factor Rev. 2004 Oct;15(5):337-51. doi: 10.1016/j.cytogfr.2004.04.001. Cytokine Growth Factor Rev. 2004. PMID: 15450250 Review.
Cited by
-
Proteolytic Cleavage of Receptor Tyrosine Kinases.Biomolecules. 2021 Apr 29;11(5):660. doi: 10.3390/biom11050660. Biomolecules. 2021. PMID: 33947097 Free PMC article. Review.
-
Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases.FEBS J. 2015 Oct;282(19):3693-721. doi: 10.1111/febs.13342. Epub 2015 Jul 4. FEBS J. 2015. PMID: 26096795 Free PMC article. Review.
-
Gamma-secretase-dependent signaling of receptor tyrosine kinases.Oncogene. 2019 Jan;38(2):151-163. doi: 10.1038/s41388-018-0465-z. Epub 2018 Aug 30. Oncogene. 2019. PMID: 30166589 Free PMC article. Review.
-
Ponatinib overcomes FGF2-mediated resistance in CML patients without kinase domain mutations.Blood. 2014 Mar 6;123(10):1516-24. doi: 10.1182/blood-2013-07-518381. Epub 2014 Jan 9. Blood. 2014. PMID: 24408322 Free PMC article.
-
VEGF-A selectively inhibits FLT1 ectodomain shedding independent of receptor activation and receptor endocytosis.Am J Physiol Cell Physiol. 2018 Aug 1;315(2):C214-C224. doi: 10.1152/ajpcell.00247.2017. Epub 2018 May 2. Am J Physiol Cell Physiol. 2018. PMID: 29719170 Free PMC article.
References
-
- Alwan HA, van Zoelen EJ, van Leeuwen JE. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J Biol Chem. 2003;278:35781–35790. - PubMed
-
- Ancot F, Foveau B, Lefebvre J, Leroy C, Tulasne D. Proteolytic cleavages give receptor tyrosine kinases the gift of ubiquity. Oncogene. 2009;28:2185–2195. - PubMed
-
- Bank U, Reinhold D, Schneemilch C, Kunz D, Synowitz HJ, Ansorge S. Selective proteolytic cleavage of IL-2 receptor and IL-6 receptor ligand binding chains by neutrophil-derived serine proteases at foci of inflammation. J Interferon Cytokine Res. 1999;19:1277–1287. - PubMed
-
- Belleudi F, Leone L, Nobili V, Raffa S, Francescangeli F, Maggio M, Morrone S, Marchese C, Torrisi MR. Keratinocyte growth factor receptor ligands target the receptor to different intracellular pathways. Traffic. 2007;8:1854–1872. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

