The functionally distinct fission yeast formins have specific actin-assembly properties
- PMID: 21865598
- PMCID: PMC3192862
- DOI: 10.1091/mbc.E11-06-0492
The functionally distinct fission yeast formins have specific actin-assembly properties
Abstract
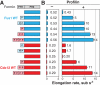
Fission yeast expresses three formins required for distinct actin cytoskeletal processes: Cdc12 (cytokinesis), For3 (polarization), and Fus1 (mating). We propose that in addition to differential regulation, key actin-assembly properties tailor formins for a particular role. In direct comparison to the well-studied Cdc12, we report the first in vitro characterization of the actin-assembly properties of For3 and Fus1. All three share fundamental formin activities; however, particular reaction rates vary significantly. Cdc12 is an efficient nucleator (one filament per approximately 3 Cdc12 dimers) that processively elongates profilin-actin at a moderate rate of 10 subunits s(-1) μM(-1), but lacks filament-bundling activity. Fus1 is also an efficient nucleator, yet processively elongates profilin-actin at one-half the rate of and dissociates 10-fold more rapidly than Cdc12; it also bundles filaments. For3 nucleates filaments 100-fold less well than Fus1, but like Cdc12, processively elongates profilin-actin at a moderate rate and lacks filament-bundling activity. Additionally, both the formin homology FH1 and FH2 domains contribute to the overall rate of profilin-actin elongation. We also confirmed the physiological importance of the actin-assembly activity of the fission yeast formins. Point mutants that disrupt their ability to stimulate actin assembly in vitro do not function properly in vivo.
Figures






Similar articles
-
The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin.J Cell Biol. 2003 Jun 9;161(5):875-87. doi: 10.1083/jcb.200211078. J Cell Biol. 2003. PMID: 12796476 Free PMC article.
-
The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis.J Cell Biol. 2013 Oct 14;203(1):101-14. doi: 10.1083/jcb.201305022. J Cell Biol. 2013. PMID: 24127216 Free PMC article.
-
Fission yeast profilin is tailored to facilitate actin assembly by the cytokinesis formin Cdc12.Mol Biol Cell. 2015 Jan 15;26(2):283-93. doi: 10.1091/mbc.E13-05-0281. Epub 2014 Nov 12. Mol Biol Cell. 2015. PMID: 25392301 Free PMC article.
-
Formins: signaling effectors for assembly and polarization of actin filaments.J Cell Sci. 2003 Jul 1;116(Pt 13):2603-11. doi: 10.1242/jcs.00611. J Cell Sci. 2003. PMID: 12775772 Review.
-
Review of the mechanism of processive actin filament elongation by formins.Cell Motil Cytoskeleton. 2009 Aug;66(8):606-17. doi: 10.1002/cm.20379. Cell Motil Cytoskeleton. 2009. PMID: 19459187 Free PMC article. Review.
Cited by
-
Cooperative bundling by fascin generates actin structures with architectures that depend on filament length.Front Cell Dev Biol. 2022 Sep 2;10:974047. doi: 10.3389/fcell.2022.974047. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36120572 Free PMC article.
-
Competition for delivery of profilin-actin to barbed ends limits the rate of formin-mediated actin filament elongation.J Biol Chem. 2020 Apr 3;295(14):4513-4525. doi: 10.1074/jbc.RA119.012000. Epub 2020 Feb 19. J Biol Chem. 2020. PMID: 32075907 Free PMC article.
-
Dynamic network morphology and tension buildup in a 3D model of cytokinetic ring assembly.Biophys J. 2014 Dec 2;107(11):2618-28. doi: 10.1016/j.bpj.2014.10.034. Epub 2014 Dec 2. Biophys J. 2014. PMID: 25468341 Free PMC article.
-
Myosin Vs organize actin cables in fission yeast.Mol Biol Cell. 2012 Dec;23(23):4579-91. doi: 10.1091/mbc.E12-07-0499. Epub 2012 Oct 10. Mol Biol Cell. 2012. PMID: 23051734 Free PMC article.
-
Negative control of cytokinesis by stress-activated MAPK signaling.Curr Genet. 2021 Oct;67(5):715-721. doi: 10.1007/s00294-021-01155-6. Epub 2021 Mar 31. Curr Genet. 2021. PMID: 33791858 Review.
References
-
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. - PubMed
-
- Castrillon DH, Wasserman SA. diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. - PubMed
-
- Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

