The mechanism of membrane-associated steps in tail-anchored protein insertion
- PMID: 21866104
- PMCID: PMC3760497
- DOI: 10.1038/nature10362
The mechanism of membrane-associated steps in tail-anchored protein insertion
Abstract
Tail-anchored (TA) membrane proteins destined for the endoplasmic reticulum are chaperoned by cytosolic targeting factors that deliver them to a membrane receptor for insertion. Although a basic framework for TA protein recognition is now emerging, the decisive targeting and membrane insertion steps are not understood. Here we reconstitute the TA protein insertion cycle with purified components, present crystal structures of key complexes between these components and perform mutational analyses based on the structures. We show that a committed targeting complex, formed by a TA protein bound to the chaperone ATPase Get3, is initially recruited to the membrane through an interaction with Get2. Once the targeting complex has been recruited, Get1 interacts with Get3 to drive TA protein release in an ATPase-dependent reaction. After releasing its TA protein cargo, the now-vacant Get3 recycles back to the cytosol concomitant with ATP binding. This work provides a detailed structural and mechanistic framework for the minimal TA protein insertion cycle.
Figures

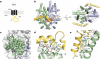
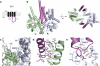


Comment in
-
It takes two to Get3.Structure. 2011 Oct 12;19(10):1353-5. doi: 10.1016/j.str.2011.10.001. Structure. 2011. PMID: 22000508
References
-
- Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. - PubMed
-
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8219–8223. - PubMed
-
- Kalbfleisch T, Cambon A, Wattenberg BW. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic. 2007;8:1687–1694. - PubMed
-
- Kriechbaumer V, et al. Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic. 2009;10:1753–1764. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

