Functional identity of the gamma tropomyosin gene: Implications for embryonic development, reproduction and cell viability
- PMID: 21866263
- PMCID: PMC3158640
- DOI: 10.4161/bioa.1.1.15172
Functional identity of the gamma tropomyosin gene: Implications for embryonic development, reproduction and cell viability
Abstract
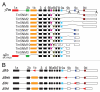
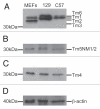
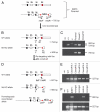
The actin filament system is fundamental to cellular functions including regulation of shape, motility, cytokinesis, intracellular trafficking and tissue organization. Tropomyosins (Tm) are highly conserved components of actin filaments which differentially regulate filament stability and function. The mammalian Tm family consists of four genes; αTm, βTm, γTm and δTm. Multiple Tm isoforms (>40) are generated by alternative splicing and expression of these isoforms is highly regulated during development. In order to further identify the role of Tm isoforms during development, we tested the specificity of function of products from the γTm gene family in mice using a series of gene knockouts. Ablation of all γTm gene cytoskeletal products results in embryonic lethality. Elimination of just two cytoskeletal products from the γTm gene (NM1,2) resulted in a 50% reduction in embryo viability. It was also not possible to generate homozygous knockout ES cells for the targets which eliminated or reduced embryo viability in mice. In contrast, homozygous knockout ES cells were generated for a different set of isoforms (NM3,5,6,8,9,11) which were not required for embryogenesis. We also observed that males hemizygous for the knockout of all cytoskeletal products from the γTm gene preferentially transmitted the minus allele with 80-100% transmission. Since all four Tm genes are expressed in early embryos, ES cells and sperm, we conclude that isoforms of the γTm gene are functionally unique in their role in embryogenesis, ES cell viability and sperm function.
Figures

References
-
- Cooper JA. Actin dynamics: tropomyosin provides stability. Curr Biol. 2002;12:523–525. - PubMed
-
- Lees-Miller JP, Helfman DM. The molecular basis for tropomyosin isoform diversity. Bioessays. 1991;13:429–437. - PubMed
-
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. - PubMed
-
- Araya E, Berthier C, Kim E, Yeung T, Wang X, Helfman DM. Regulation of coiled-coil assembly in tropomyosins. J Struct Biol. 2002;137:176–183. - PubMed
-
- Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
