A multi-organ transcriptome resource for the Burmese Python (Python molurus bivittatus)
- PMID: 21867488
- PMCID: PMC3173347
- DOI: 10.1186/1756-0500-4-310
A multi-organ transcriptome resource for the Burmese Python (Python molurus bivittatus)
Abstract
Background: Snakes provide a unique vertebrate system for studying a diversity of extreme adaptations, including those related to development, metabolism, physiology, and venom. Despite their importance as research models, genomic resources for snakes are few. Among snakes, the Burmese python is the premier model for studying extremes of metabolic fluctuation and physiological remodelling. In this species, the consumption of large infrequent meals can induce a 40-fold increase in metabolic rate and more than a doubling in size of some organs. To provide a foundation for research utilizing the python, our aim was to assemble and annotate a transcriptome reference from the heart and liver. To accomplish this aim, we used the 454-FLX sequencing platform to collect sequence data from multiple cDNA libraries.
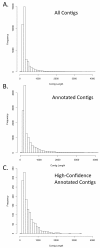
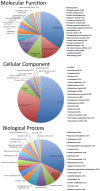
Results: We collected nearly 1 million 454 sequence reads, and assembled these into 37,245 contigs with a combined length of 13,409,006 bp. To identify known genes, these contigs were compared to chicken and lizard gene sets, and to all Genbank sequences. A total of 13,286 of these contigs were annotated based on similarity to known genes or Genbank sequences. We used gene ontology (GO) assignments to characterize the types of genes in this transcriptome resource. The raw data, transcript contig assembly, and transcript annotations are made available online for use by the broader research community.
Conclusion: These data should facilitate future studies using pythons and snakes in general, helping to further contribute to the utilization of snakes as a model evolutionary and physiological system. This sequence collection represents a major genomic resource for the Burmese python, and the large number of transcript sequences characterized should contribute to future research in this and other snake species.
Figures


References
-
- Ott BD, Secor SM. Adaptive regulation of digestive performance in the genus Python. J Exper Biol. 2007;210(Pt 2):340–356. - PubMed
LinkOut - more resources
Full Text Sources

