Analysis of human meiotic recombination events with a parent-sibling tracing approach
- PMID: 21867557
- PMCID: PMC3186786
- DOI: 10.1186/1471-2164-12-434
Analysis of human meiotic recombination events with a parent-sibling tracing approach
Abstract
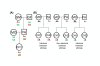
Background: Meiotic recombination ensures that each child inherits distinct genetic materials from each parent, but the distribution of crossovers along meiotic chromosomes remains difficult to identify. In this study, we developed a parent-sibling tracing (PST) approach from previously reported methods to identify meiotic crossover sites of GEO GSE6754 data set. This approach requires only the single nucleotide polymorphism (SNP) data of the pedigrees of both parents and at least two of children.
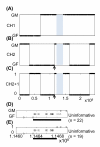
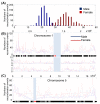
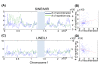
Results: Compared to other SNP-based algorithms (identity by descent or pediSNP), fewer uninformative SNPs were derived with the use of PST. Analysis of a GEO GSE6754 data set containing 2,145 maternal and paternal meiotic events revealed that the pattern and distribution of paternal and maternal recombination sites vary along the chromosomes. Lower crossover rates near the centromeres were more prominent in males than in females. Based on analysis of repetitive sequences, we also showed that recombination hotspots are positively correlated with SINE/MIR repetitive elements and negatively correlated with LINE/L1 elements. The number of meiotic recombination events was positively correlated with the number of shorter tandem repeat sequences.
Conclusions: The advantages of the PST approach include the ability to use only two-generation pedigrees with two siblings and the ability to perform gender-specific analyses of repetitive elements and tandem repeat sequences while including fewer uninformative SNP regions in the results.
Figures
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of THE CELL. 5. New York: Garland Science; 2008.
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

