CD8α(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes
- PMID: 21867927
- PMCID: PMC3172670
- DOI: 10.1016/j.immuni.2011.06.012
CD8α(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes
Abstract
CD8α(+) dendritic cells (DCs) prime cytotoxic T lymphocytes during viral infections and produce interleukin-12 in response to pathogens. Although the loss of CD8α(+) DCs in Batf3(-/-) mice increases their susceptibility to several pathogens, we observed that Batf3(-/-) mice exhibited enhanced resistance to the intracellular bacterium Listeria monocytogenes. In wild-type mice, Listeria organisms, initially located in the splenic marginal zone, migrated to the periarteriolar lymphoid sheath (PALS) where they grew exponentially and induced widespread lymphocyte apoptosis. In Batf3(-/-) mice, however, Listeria organisms remain trapped in the marginal zone, failed to traffic into the PALS, and were rapidly cleared by phagocytes. In addition, Batf3(-/-) mice, which lacked the normal population of hepatic CD103(+) peripheral DCs, also showed protection from liver infection. These results suggest that Batf3-dependent CD8α(+) and CD103(+) DCs provide initial cellular entry points within the reticuloendothelial system by which Listeria establishes productive infection.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
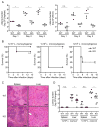

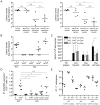
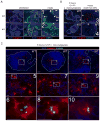
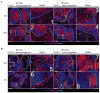


Comment in
-
Intracellular pathogens and CD8(+) dendritic cells: dangerous liaisons.Immunity. 2011 Aug 26;35(2):153-5. doi: 10.1016/j.immuni.2011.08.003. Immunity. 2011. PMID: 21867923
References
-
- Alaniz RC, Sandall S, Thomas EK, Wilson CB. Increased dendritic cell numbers impair protective immunity to intracellular bacteria despite augmenting antigen-specific CD8+ T lymphocyte responses. J Immunol. 2004;172:3725–3735. - PubMed
-
- Aliberti J, Schulz O, Pennington DJ, Tsujimura H, Sousa Reis E, Ozato K, Sher A. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. - PubMed
-
- Aoshi T, Zinselmeyer BH, Konjufca V, Lynch JN, Zhang X, Koide Y, Miller MJ. Bacterial entry to the splenic white pulp initiates antigen presentation to CD8+ T cells. Immunity. 2008;29:476–486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

