Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: modelling the ligand-bound receptor
- PMID: 21868452
- PMCID: PMC5417182
- DOI: 10.1210/me.2011-1160
Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: modelling the ligand-bound receptor
Abstract
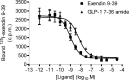
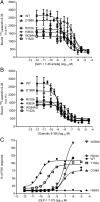
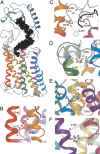
The C-terminal regions of glucagon-like peptide-1 (GLP-1) bind to the N terminus of the GLP-1 receptor (GLP-1R), facilitating interaction of the ligand N terminus with the receptor transmembrane domain. In contrast, the agonist exendin-4 relies less on the transmembrane domain, and truncated antagonist analogs (e.g. exendin 9-39) may interact solely with the receptor N terminus. Here we used mutagenesis to explore the role of residues highly conserved in the predicted transmembrane helices of mammalian GLP-1Rs and conserved in family B G protein coupled receptors in ligand binding and GLP-1R activation. By iteration using information from the mutagenesis, along with the available crystal structure of the receptor N terminus and a model of the active opsin transmembrane domain, we developed a structural receptor model with GLP-1 bound and used this to better understand consequences of mutations. Mutation at Y152 [transmembrane helix (TM) 1], R190 (TM2), Y235 (TM3), H363 (TM6), and E364 (TM6) produced similar reductions in affinity for GLP-1 and exendin 9-39. In contrast, other mutations either preferentially [K197 (TM2), Q234 (TM3), and W284 (extracellular loop 2)] or solely [D198 (TM2) and R310 (TM5)] reduced GLP-1 affinity. Reduced agonist affinity was always associated with reduced potency. However, reductions in potency exceeded reductions in agonist affinity for K197A, W284A, and R310A, while H363A was uncoupled from cAMP generation, highlighting critical roles of these residues in translating binding to activation. Data show important roles in ligand binding and receptor activation of conserved residues within the transmembrane domain of the GLP-1R. The receptor structural model provides insight into the roles of these residues.
Figures

References
-
- Orskov C , Rabenhøj L , Wettergren A , Kofod H , Holst JJ. 1994. Tissue and plasma concentrations of amidated and glycine- extended glucagon-like peptide I in humans. Diabetes 43:535–539 - PubMed
-
- Bavec A , Hällbrink M , Langel U , Zorko M. 2003. Different role of intracellular loops of glucagon-like peptide-1 receptor in G-protein coupling. Regul Pept 111:137–144 - PubMed
-
- Hällbrink M , Holmqvist T , Olsson M , Ostenson CG , Efendic S , Langel U. 2001. Different domains in the third intracellular loop of the GLP-1 receptor are responsible for Gαs and Gαi/Gαo activation. Biochim Biophys Acta 1546:79–86 - PubMed
-
- Montrose-Rafizadeh C , Avdonin P , Garant MJ , Rodgers BD , Kole S , Yang H , Levine MA , Schwindinger W , Bernier M. 1999. Pancreatic glucagon-like peptide-1 receptor couples to multiple G proteins and activates mitogen-activated protein kinase pathways in Chinese hamster ovary cells. Endocrinology 140:1132–1140 - PubMed
-
- Baggio LL , Drucker DJ. 2007. Biology of incretins: GLP-1 and GIP. Gastroenterology 132:2131–2157 - PubMed

