Complex mechanisms regulate developmental expression of the matA (HMG) mating type gene in homothallic Aspergillus nidulans
- PMID: 21868608
- PMCID: PMC3213371
- DOI: 10.1534/genetics.111.131458
Complex mechanisms regulate developmental expression of the matA (HMG) mating type gene in homothallic Aspergillus nidulans
Abstract
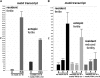
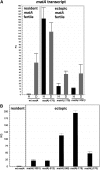
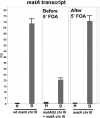
Sexual reproduction is a fundamental developmental process that allows for genetic diversity through the control of zygote formation, recombination, and gametogenesis. The correct regulation of these events is paramount. Sexual reproduction in filamentous fungi, including mating strategy (self-fertilization/homothallism or outcrossing/heterothallism), is determined by the expression of mating type genes at mat loci. Aspergillus nidulans matA encodes a critical regulator that is a fungal ortholog of the hSRY/SOX9 HMG box proteins. In contrast to well-studied outcrossing systems, the molecular basis of homothallism and role of mating type genes during a self-fertile sexual cycle remain largely unknown. In this study the genetic model organism, A. nidulans, has been used to investigate the regulation and molecular functions of the matA mating type gene in a homothallic system. Our data demonstrate that complex regulatory mechanisms underlie functional matA expression during self-fertilization and sexual reproduction in A. nidulans. matA expression is suppressed in vegetative hyphae and is progressively derepressed during the sexual cycle. Elevated levels of matA transcript are required for differentiation of fruiting bodies, karyogamy, meiosis, and efficient formation of meiotic progeny. matA expression is driven from both initiator (Inr) and novel promoter elements that are tightly developmentally regulated by position-dependent and position-independent mechanisms. Deletion of an upstream silencing element, matA SE, results in derepressed expression from wild-type (wt) promoter elements and activation of an additional promoter. These studies provide novel insights into the molecular basis of homothallism in fungi and genetic regulation of sexual reproduction in eukaryotes.
Figures






Similar articles
-
Novel sexual-cycle-specific gene silencing in Aspergillus nidulans.Genetics. 2013 Apr;193(4):1149-62. doi: 10.1534/genetics.112.147546. Epub 2013 Jan 22. Genetics. 2013. PMID: 23341415 Free PMC article.
-
Mating type protein Mat1-2 from asexual Aspergillus fumigatus drives sexual reproduction in fertile Aspergillus nidulans.Eukaryot Cell. 2008 Jun;7(6):1029-40. doi: 10.1128/EC.00380-07. Epub 2008 Feb 1. Eukaryot Cell. 2008. PMID: 18245277 Free PMC article.
-
Structural and functional conservation of fungal MatA and human SRY sex-determining proteins.Nat Commun. 2014 Nov 17;5:5434. doi: 10.1038/ncomms6434. Nat Commun. 2014. PMID: 25399555 Free PMC article.
-
An Overview of the Function and Maintenance of Sexual Reproduction in Dikaryotic Fungi.Front Microbiol. 2018 Mar 21;9:503. doi: 10.3389/fmicb.2018.00503. eCollection 2018. Front Microbiol. 2018. PMID: 29619017 Free PMC article. Review.
-
Genetic Networks That Govern Sexual Reproduction in the Pezizomycotina.Microbiol Mol Biol Rev. 2021 Dec 15;85(4):e0002021. doi: 10.1128/MMBR.00020-21. Epub 2021 Sep 29. Microbiol Mol Biol Rev. 2021. PMID: 34585983 Free PMC article. Review.
Cited by
-
The Role of Chromatin and Transcriptional Control in the Formation of Sexual Fruiting Bodies in Fungi.Microbiol Mol Biol Rev. 2022 Dec 21;86(4):e0010422. doi: 10.1128/mmbr.00104-22. Epub 2022 Nov 21. Microbiol Mol Biol Rev. 2022. PMID: 36409109 Free PMC article. Review.
-
Discovery of New Genomic Configuration of Mating-Type Loci in the Largest Lineage of Lichen-Forming Fungi.Genome Biol Evol. 2024 May 2;16(5):evae094. doi: 10.1093/gbe/evae094. Genome Biol Evol. 2024. PMID: 38686438 Free PMC article.
-
Aspergillus: sex and recombination.Mycopathologia. 2014 Dec;178(5-6):349-62. doi: 10.1007/s11046-014-9795-8. Epub 2014 Aug 14. Mycopathologia. 2014. PMID: 25118872 Review.
-
Genetic Dissection of Sexual Reproduction in a Primary Homothallic Basidiomycete.PLoS Genet. 2016 Jun 21;12(6):e1006110. doi: 10.1371/journal.pgen.1006110. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27327578 Free PMC article.
-
Novel sexual-cycle-specific gene silencing in Aspergillus nidulans.Genetics. 2013 Apr;193(4):1149-62. doi: 10.1534/genetics.112.147546. Epub 2013 Jan 22. Genetics. 2013. PMID: 23341415 Free PMC article.
References
-
- Benjamin C. R., 1955. Ascocarps of Aspergillus and Penicillium, pp. 669–687 in Mycologia. Mycological Society of America, Washington, DC
-
- Boeke J. D., LaCroute F., Fink G. R., 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197: 345–346 - PubMed
-
- Champe S. P., Nagle D. L., Yager L. N., 1994. Sexual sporulation. Prog. Ind. Microbiol. 29: 429–454 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

