The shaping of modern human immune systems by multiregional admixture with archaic humans
- PMID: 21868630
- PMCID: PMC3677943
- DOI: 10.1126/science.1209202
The shaping of modern human immune systems by multiregional admixture with archaic humans
Abstract
Whole genome comparisons identified introgression from archaic to modern humans. Our analysis of highly polymorphic human leukocyte antigen (HLA) class I, vital immune system components subject to strong balancing selection, shows how modern humans acquired the HLA-B*73 allele in west Asia through admixture with archaic humans called Denisovans, a likely sister group to the Neandertals. Virtual genotyping of Denisovan and Neandertal genomes identified archaic HLA haplotypes carrying functionally distinctive alleles that have introgressed into modern Eurasian and Oceanian populations. These alleles, of which several encode unique or strong ligands for natural killer cell receptors, now represent more than half the HLA alleles of modern Eurasians and also appear to have been later introduced into Africans. Thus, adaptive introgression of archaic alleles has significantly shaped modern human immune systems.
Figures

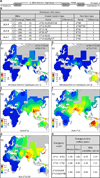
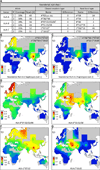

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

