Relative Expression Levels Rather Than Specific Activity Plays the Major Role in Determining In Vivo AKT Isoform Substrate Specificity
- PMID: 21869924
- PMCID: PMC3160084
- DOI: 10.4061/2011/720985
Relative Expression Levels Rather Than Specific Activity Plays the Major Role in Determining In Vivo AKT Isoform Substrate Specificity
Abstract
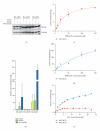
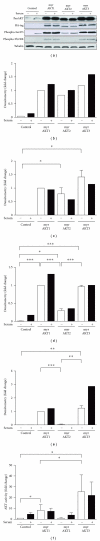
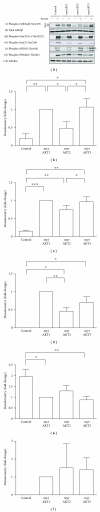
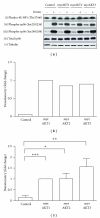
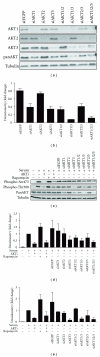
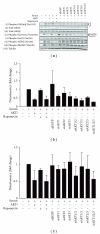
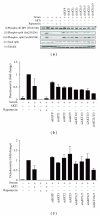
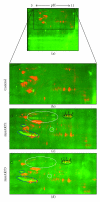
The AKT protooncogene mediates many cellular processes involved in normal development and disease states such as cancer. The three structurally similar isoforms: AKT1, AKT2, and AKT3 exhibit both functional redundancy and isoform-specific functions; however the basis for their differential signalling remains unclear. Here we show that in vitro, purified AKT3 is ∼47-fold more active than AKT1 at phosphorylating peptide and protein substrates. Despite these marked variations in specific activity between the individual isoforms, a comprehensive analysis of phosphorylation of validated AKT substrates indicated only subtle differences in signalling via individual isoforms in vivo. Therefore, we hypothesise, at least in this model system, that relative tissue/cellular abundance, rather than specific activity, plays the dominant role in determining AKT substrate specificity in situ.
Figures
References
-
- Kumar CC, Madison V. AKT crystal structure and AKT-specific inhibitors. Oncogene. 2005;24(50):7493–7501. - PubMed
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. - PubMed
-
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nature Reviews Cancer. 2002;2(7):489–501. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

