Dramatic increase in the signal and sensitivity of detection via self-assembly of branched DNA
- PMID: 21870112
- PMCID: PMC3887650
- DOI: 10.1007/s10059-011-0121-8
Dramatic increase in the signal and sensitivity of detection via self-assembly of branched DNA
Abstract
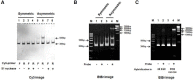
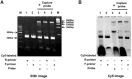
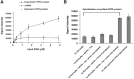
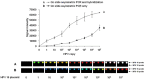
In molecular testing using PCR, the target DNA is amplified via PCR and the sequence of interest is investigated via hybridization with short oligonucleotide capture probes that are either in a solution or immobilized on solid supports such as beads or glass slides. In this report, we report the discovery of assembly of DNA complex(es) between a capture probe and multiple strands of the PCR product. The DNA complex most likely has branched structure. The assembly of branched DNA was facilitated by the product of asymmetric PCR. The amount of branched DNA assembled was increased five fold when the asymmetric PCR product was denatured and hybridized with a capture probe all in the same PCR reaction mixture. The major branched DNA species appeared to contain three reverse strands (the strand complementary to the capture probe) and two forward strands. The DNA was sensitive to S1 nuclease suggesting that it had single-stranded gaps. Branched DNA also appeared to be assembled with the capture probes immobilized on the surface of solid support when the product of asymmetric PCR was hybridized. Assembly of the branched DNA was also increased when hybridization was performed in complete PCR reaction mixture suggesting the requirement of DNA synthesis. Integration of asymmetric PCR, heat denaturation and hybridization in the same PCR reaction mixture with the capture probes immobilized on the surface of solid support achieved dramatic increase in the signal and sensitivity of detection of DNA. Such a system should be advantageously applied for development of automated process for detection of DNA.
Figures


Similar articles
-
Hybridization of glass-tethered oligonucleotide probes to target strands preannealed with labeled auxiliary oligonucleotides.Mol Biotechnol. 1999 Feb;11(1):1-12. doi: 10.1007/BF02789172. Mol Biotechnol. 1999. PMID: 10367278
-
Electrochemical genosensor based on peptide nucleic acid-mediated PCR and asymmetric PCR techniques: Electrostatic interactions with a metal cation.Anal Chem. 2006 Apr 1;78(7):2182-9. doi: 10.1021/ac051526a. Anal Chem. 2006. PMID: 16579596
-
A nuclease-polymerase chain reaction enables amplification of probes used for capture-based DNA target enrichment.Nucleic Acids Res. 2019 Dec 16;47(22):e147. doi: 10.1093/nar/gkz870. Nucleic Acids Res. 2019. PMID: 31598677 Free PMC article.
-
Correlation between microarray DNA hybridization efficiency and the position of short capture probe on the target nucleic acid.Biotechniques. 2005 Jul;39(1):89-96. doi: 10.2144/05391RR01. Biotechniques. 2005. PMID: 16060373
-
Colorimetric detection for PCR amplified HIV-1 DNA using magnetic beads.J Virol Methods. 1993 Mar;41(3):341-50. doi: 10.1016/0166-0934(93)90023-k. J Virol Methods. 1993. PMID: 8473372
Cited by
-
Single-stranded DNA catalyzes hybridization of PCR-products to microarray capture probes.PLoS One. 2014 Jul 15;9(7):e102338. doi: 10.1371/journal.pone.0102338. eCollection 2014. PLoS One. 2014. PMID: 25025686 Free PMC article.
References
-
- Albrecht V., Chevallier A., Magnone V., Barbry P., Vandenbos F., Bongain A., Lefebvre J.C., Giordanengo V. Easy and fast detection and genotyping of high-risk human papillomaviruse by dedicated DNA microarrays. J. Virol. Methods. (2006);137:236–244. - PubMed
-
- Brewer B.J., Fangman W.L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. (1987);56:463–471. - PubMed
-
- Chung I.H., Yoo H.S., Eah J.Y., Yoon H.K., Jung J.W., Hwang S.Y., Kim C.B. A DNA microarray for idenficiation of seleceted Korean birds based on mitochondrial cytochrome c oxidase I gene sequences. Mol. Cells. (2010);30:295–301. - PubMed
-
- Dunderdale H.J., Benson F.E., Parson C.A., Sharples G.J., Lloyd R.G., West S.G. Formation and resolution of recombination intermediates by E.coli RecA and RuvC proteins. Nature. (1991);354:506–510. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

