Promiscuous partnerships in Ewing's sarcoma
- PMID: 21872822
- PMCID: PMC3164520
- DOI: 10.1016/j.cancergen.2011.07.008
Promiscuous partnerships in Ewing's sarcoma
Abstract
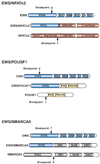
Ewing's sarcoma is a highly aggressive bone and soft tissue tumor of children and young adults. At the molecular genetic level Ewing's sarcoma is characterized by a balanced reciprocal translocation, t(11;22)(q24;q12), which encodes an oncogenic fusion protein and transcription factor EWS/FLI. This tumor-specific chimeric fusion retains the amino terminus of EWS, a member of the TET (TLS/EWS/TAF15) family of RNA-binding proteins, and the carboxy terminus of FLI, a member of the ETS family of transcription factors. In addition to EWS/FLI, variant translocation fusions belonging to the TET/ETS family have been identified in Ewing's sarcoma. These studies solidified the importance of TET/ETS fusions in the pathogenesis of Ewing's sarcoma and have since been used as diagnostic markers for the disease. EWS fusions with non-ETS transcription factor family members have been described in sarcomas that are clearly distinct from Ewing's sarcoma. However, in recent years there have been reports of rare fusions in "Ewing's-like tumors" that harbor the amino-terminus of EWS fused to the carboxy-terminal DNA or chromatin-interacting domains contributed by non-ETS proteins. This review aims to summarize the growing list of fusion oncogenes that characterize Ewing's sarcoma and Ewing's-like tumors and highlights important questions that need to be answered to further support the existing concept that Ewing's sarcoma is strictly a "TET/ETS" fusion-driven malignancy. Understanding the molecular mechanisms of action of the various different fusion oncogenes will provide better insights into the biology underlying this rare but important solid tumor.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



References
-
- Janknecht R. EWS-ETS oncoproteins: the linchpins of Ewing tumors. Gene. 2005;363:1–14. - PubMed
-
- Ewing J. Diffuse Endothelioma of Bone. Proceedings of the New York Pathological Society. 1921;21:17–24. - PubMed
-
- Stiller CA, Bielack SS, Jundt G, et al. Bone tumours in European children and adolescents, 1978–1997. Report from the Automated Childhood Cancer Information System project. Eur J Cancer. 2006;42:2124–2135. - PubMed
-
- Denny CT. Ewing's sarcoma--a clinical enigma coming into focus. J Pediatr Hematol Oncol. 1998;20:421–425. - PubMed
-
- Paulussen M, Frohlich B, Jurgens H. Ewing tumour: incidence, prognosis and treatment options. Paediatr Drugs. 2001;3:899–913. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

