Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing
- PMID: 21873192
- PMCID: PMC3169109
- DOI: 10.1073/pnas.1112213108
Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing
Abstract
Characterizing the genetic programs that specify development and evolution of the cerebral cortex is a central challenge in neuroscience. Stem cells in the transient embryonic ventricular and subventricular zones generate neurons that migrate across the intermediate zone to the overlying cortical plate, where they differentiate and form the neocortex. It is clear that not one but a multitude of molecular pathways are necessary to progress through each cellular milestone, yet the underlying transcriptional programs remain unknown. Here, we apply differential transcriptome analysis on microscopically isolated cell populations, to define five transcriptional programs that represent each transient embryonic zone and the progression between these zones. The five transcriptional programs contain largely uncharacterized genes in addition to transcripts necessary for stem cell maintenance, neurogenesis, migration, and differentiation. Additionally, we found intergenic transcriptionally active regions that possibly encode unique zone-specific transcripts. Finally, we present a high-resolution transcriptome map of transient zones in the embryonic mouse forebrain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

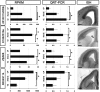

References
-
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. - PubMed
-
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. - PubMed
-
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

