Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk
- PMID: 21873198
- PMCID: PMC3169104
- DOI: 10.1073/pnas.1111691108
Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk
Abstract
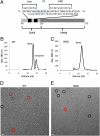
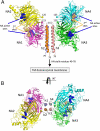
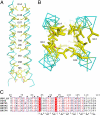
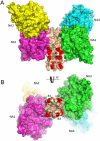
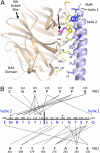
The paramyxovirus hemagglutinin-neuraminidase (HN) protein plays multiple roles in viral entry and egress, including binding to sialic acid receptors, activating the fusion (F) protein to activate membrane fusion and viral entry, and cleaving sialic acid from carbohydrate chains. HN is an oligomeric integral membrane protein consisting of an N-terminal transmembrane domain, a stalk region, and an enzymatically active neuraminidase (NA) domain. Structures of the HN NA domains have been solved previously; however, the structure of the stalk region has remained elusive. The stalk region contains specificity determinants for F interactions and activation, underlying the requirement for homotypic F and HN interactions in viral entry. Mutations of the Newcastle disease virus HN stalk region have been shown to affect both F activation and NA activities, but a structural basis for understanding these dual affects on HN functions has been lacking. Here, we report the structure of the Newcastle disease virus HN ectodomain, revealing dimers of NA domain dimers flanking the N-terminal stalk domain. The stalk forms a parallel tetrameric coiled-coil bundle (4HB) that allows classification of extensive mutational data, providing insight into the functional roles of the stalk region. Mutations that affect both F activation and NA activities map predominantly to the 4HB hydrophobic core, whereas mutations that affect only F-protein activation map primarily to the 4HB surface. Two of four NA domains interact with the 4HB stalk, and residues at this interface in both the stalk and NA domain have been implicated in HN function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1449–1496.
-
- Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. - PubMed
-
- Takimoto T, Taylor GL, Crennell SJ, Scroggs RA, Portner A. Crystallization of Newcastle disease virus hemagglutinin-neuraminidase glycoprotein. Virology. 2000;270:208–214. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

