Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis
- PMID: 21873203
- PMCID: PMC3169142
- DOI: 10.1073/pnas.1111577108
Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis
Abstract
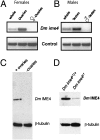
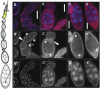
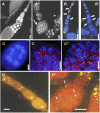
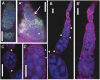
N(6)-methyladenosine is a nonediting RNA modification found in mRNA of all eukaryotes, from yeast to humans. Although the functional significance of N(6)-methyladenosine is unknown, the Inducer of MEiosis 4 (IME4) gene of Saccharomyces cerevisiae, which encodes the enzyme that catalyzes this modification, is required for gametogenesis. Here we find that the Drosophila IME4 homolog, Dm ime4, is expressed in ovaries and testes, indicating an evolutionarily conserved function for this enzyme in gametogenesis. In contrast to yeast, but as in Arabidopsis, Dm ime4 is essential for viability. Lethality is rescued fully by a wild-type transgenic copy of Dm ime4 but not by introducing mutations shown to abrogate the catalytic activity of yeast Ime4, indicating functional conservation of the catalytic domain. The phenotypes of hypomorphic alleles of Dm ime4 that allow recovery of viable adults reveal critical functions for this gene in oogenesis. Ovarioles from Dm ime4 mutants have fused egg chambers with follicle-cell defects similar to those observed when Notch signaling is defective. Indeed, using a reporter for Notch activation, we find markedly reduced levels of Notch signaling in follicle cells of Dm ime4 mutants. This phenotype of Dm ime4 mutants is rescued by inducing expression of a constitutively activated form of Notch. Our study reveals the function of IME4 in a metazoan. In yeast, this enzyme is responsible for a crucial developmental decision, whereas in Drosophila it appears to target the conserved Notch signaling pathway, which regulates many vital aspects of metazoan development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

