Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa
- PMID: 21873404
- PMCID: PMC3352280
- DOI: 10.1099/mic.0.051987-0
Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa
Abstract
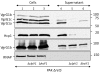
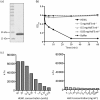
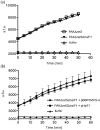
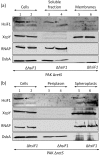
Bacterial pathogens use a range of protein secretion systems to colonize their host. One recent addition to this arsenal is the type VI secretion system (T6SS), which is found in many Gram-negative bacteria. The T6SS involves 12-15 components, including a ClpV-like AAA(+) ATPase. Moreover, the VgrG and Hcp components have been proposed to form a puncturing device, based on structural similarity to the tail spike components gp5/gp27 and the tail tube component gp19 of the T4 bacteriophage, respectively. Another T6SS component shows similarity to a T4 phage protein, namely gp25. The gp25 protein has been proposed to have lysozyme activity. Other T6SS components do not exhibit obvious similarity to characterized T4 phage components. The genome of Pseudomonas aeruginosa contains three T6SS gene clusters. In each cluster a gene encoding a putative member of the gp25-like protein family was identified, which we called HsiF. We confirmed this similarity by analysing the structure of the P. aeruginosa HsiF proteins using secondary and tertiary structure prediction tools. We demonstrated that HsiF1 is crucial for the T6SS-dependent secretion of Hcp and VgrG. Importantly, lysozyme activity of HsiF proteins was not detectable, and we related this observation to the demonstration that HsiF1 localizes to the cytoplasm of P. aeruginosa. Finally, our data showed that a conserved glutamate, predicted to be required for proper HsiF folding, is essential for its function. In conclusion, our data confirm the central role of HsiF in the T6SS mechanism, provide information on the predicted HsiF structure, and call for reconsideration of the function of gp25-like proteins.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

