Inactive-state preassembly of G(q)-coupled receptors and G(q) heterotrimers
- PMID: 21873996
- PMCID: PMC3177959
- DOI: 10.1038/nchembio.642
Inactive-state preassembly of G(q)-coupled receptors and G(q) heterotrimers
Abstract
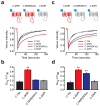
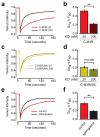
G protein-coupled receptors (GPCRs) transmit signals by forming active-state complexes with heterotrimeric G proteins. It has been suggested that some GPCRs also assemble with G proteins before ligand-induced activation and that inactive-state preassembly facilitates rapid and specific G protein activation. However, no mechanism of preassembly has been described, and no functional consequences of preassembly have been demonstrated. Here we show that M(3) muscarinic acetylcholine receptors (M3R) form inactive-state complexes with G(q) heterotrimers in intact cells. The M3R C terminus is sufficient, and a six-amino-acid polybasic sequence distal to helix 8 ((565)KKKRRK(570)) is necessary for preassembly with G(q). Replacing this sequence with six alanine residues prevents preassembly, slows the rate of G(q) activation and decreases steady-state agonist sensitivity. That other G(q)-coupled receptors possess similar polybasic regions and also preassemble with G(q) suggests that these GPCRs may use a common preassembly mechanism to facilitate activation of G(q) heterotrimers.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Comment in
-
Receptors: GPCR-G protein preassembly?Nat Chem Biol. 2011 Sep 19;7(10):657-8. doi: 10.1038/nchembio.665. Nat Chem Biol. 2011. PMID: 21931312 No abstract available.
References
-
- Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. - PubMed
-
- De Lean A, Stadel JM, Lefkowitz RJ. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J Biol Chem. 1980;255:7108–7117. - PubMed
-
- Oldham WM, Hamm HE. Structural basis of function in heterotrimeric G proteins. Q Rev Biophys. 2006;39:117–166. - PubMed
-
- Wess J, et al. Structural basis of receptor/G protein coupling selectivity studied with muscarinic receptors as model systems. Life Sci. 1997;60:1007–1014. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

