Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase
- PMID: 21874008
- PMCID: PMC3186838
- DOI: 10.1038/ncb2327
Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase
Abstract
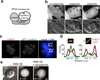
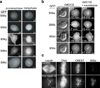
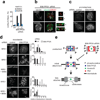
Error-free chromosome segregation depends on the precise regulation of phosphorylation to stabilize kinetochore-microtubule attachments (K-fibres) on sister chromatids that have attached to opposite spindle poles (bi-oriented). In many instances, phosphorylation correlates with K-fibre destabilization. Consistent with this, multiple kinases, including Aurora B and Plk1, are enriched at kinetochores of mal-oriented chromosomes when compared with bi-oriented chromosomes, which have stable attachments. Paradoxically, however, these kinases also target to prometaphase chromosomes that have not yet established spindle attachments and it is therefore unclear how kinetochore-microtubule interactions can be stabilized when kinase levels are high. Here we show that the generation of stable K-fibres depends on the B56-PP2A phosphatase, which is enriched at centromeres/kinetochores of unattached chromosomes. When B56-PP2A is depleted, K-fibres are destabilized and chromosomes fail to align at the spindle equator. Strikingly, B56-PP2A depletion increases the level of phosphorylation of Aurora B and Plk1 kinetochore substrates as well as Plk1 recruitment to kinetochores. Consistent with increased substrate phosphorylation, we find that chemical inhibition of Aurora or Plk1 restores K-fibres in B56-PP2A-depleted cells. Our findings reveal that PP2A, an essential tumour suppressor, tunes the balance of phosphorylation to promote chromosome-spindle interactions during cell division.
Figures


References
-
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. - PubMed
-
- Ahonen LJ, et al. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr Biol. 2005;15:1078–1089. - PubMed
-
- Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. - PubMed
-
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. - PubMed
-
- Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. - PubMed
Materials and Methods References
-
- Tang Z, et al. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

