In vitro centromere and kinetochore assembly on defined chromatin templates
- PMID: 21874020
- PMCID: PMC3175311
- DOI: 10.1038/nature10379
In vitro centromere and kinetochore assembly on defined chromatin templates
Abstract
During cell division, chromosomes are segregated to nascent daughter cells by attaching to the microtubules of the mitotic spindle through the kinetochore. Kinetochores are assembled on a specialized chromatin domain called the centromere, which is characterized by the replacement of nucleosomal histone H3 with the histone H3 variant centromere protein A (CENP-A). CENP-A is essential for centromere and kinetochore formation in all eukaryotes but it is unknown how CENP-A chromatin directs centromere and kinetochore assembly. Here we generate synthetic CENP-A chromatin that recapitulates essential steps of centromere and kinetochore assembly in vitro. We show that reconstituted CENP-A chromatin when added to cell-free extracts is sufficient for the assembly of centromere and kinetochore proteins, microtubule binding and stabilization, and mitotic checkpoint function. Using chromatin assembled from histone H3/CENP-A chimaeras, we demonstrate that the conserved carboxy terminus of CENP-A is necessary and sufficient for centromere and kinetochore protein recruitment and function but that the CENP-A targeting domain--required for new CENP-A histone assembly--is not. These data show that two of the primary requirements for accurate chromosome segregation, the assembly of the kinetochore and the propagation of CENP-A chromatin, are specified by different elements in the CENP-A histone. Our unique cell-free system enables complete control and manipulation of the chromatin substrate and thus presents a powerful tool to study centromere and kinetochore assembly.
Figures
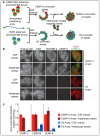
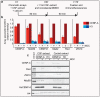
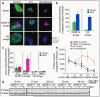
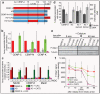
Comment in
-
Cell division: Six degrees of separation.Nature. 2011 Sep 14;477(7364):283-5. doi: 10.1038/477283a. Nature. 2011. PMID: 21921905 No abstract available.
-
Cell division: CENPA's tail rules the centromere.Nat Rev Mol Cell Biol. 2011 Sep 23;12(10):626. doi: 10.1038/nrm3196. Nat Rev Mol Cell Biol. 2011. PMID: 21941272 No abstract available.
References
-
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. - PubMed
-
- Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. - PubMed
-
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

