Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome
- PMID: 21875594
- PMCID: PMC4451184
- DOI: 10.1016/j.jmb.2011.08.031
Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome
Abstract
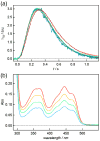
Cryptochromes (CRYs) are widespread flavoproteins with homology to photolyases (PHRs), a class of blue-light-activated DNA repair enzymes. Unlike PHRs, both plant and animal CRYs have a C-terminal domain. This cryptochrome C-terminal (CCT) domain mediates interactions with other proteins, while the PHR-like domain converts light energy into a signal via reduction and radical formation of the flavin adenine dinucleotide cofactor. However, the mechanism by which the PHR-like domain regulates the CCT domain is not known. Here, we applied the pulsed-laser-induced transient grating method to detect conformational changes induced by blue-light excitation of full-length Arabidopsis thaliana cryptochrome 1 (AtCRY1). A significant reduction in the diffusion coefficient of AtCRY1 was observed upon photoexcitation, indicating that a large conformational change occurs in this monomeric protein. AtCRY1 containing a single mutation (W324F) that abolishes an intra-protein electron transfer cascade did not exhibit this conformational change. Moreover, the conformational change was much reduced in protein lacking the CCT domain. Thus, we conclude that the observed large conformational changes triggered by light excitation of the PHR-like domain result from C-terminal domain rearrangement. This inter-domain modulation would be critical for CRYs' ability to transduce a blue-light signal into altered protein-protein interactions for biological activity. Lastly, we demonstrate that the transient grating technique provides a powerful method for the direct observation and understanding of photoreceptor dynamics.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function.J Biol Chem. 2005 May 20;280(20):19437-40. doi: 10.1074/jbc.C500077200. Epub 2005 Mar 17. J Biol Chem. 2005. PMID: 15774475
-
Substitution of a conserved glycine in the PHR domain of Arabidopsis cryptochrome 1 confers a constitutive light response.Mol Plant. 2012 Jan;5(1):85-97. doi: 10.1093/mp/ssr052. Epub 2011 Jul 16. Mol Plant. 2012. PMID: 21765176
-
Cellular metabolites modulate in vivo signaling of Arabidopsis cryptochrome-1.Plant Signal Behav. 2015;10(9):e1063758. doi: 10.1080/15592324.2015.1063758. Plant Signal Behav. 2015. PMID: 26313597 Free PMC article.
-
Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana.J Plant Res. 2016 Mar;129(2):137-48. doi: 10.1007/s10265-015-0782-z. Epub 2016 Jan 25. J Plant Res. 2016. PMID: 26810763 Free PMC article. Review.
-
Conformational and Intermolecular Interaction Dynamics of Photolyase/Cryptochrome Proteins Monitored by the Time-Resolved Diffusion Technique.Photochem Photobiol. 2017 Jan;93(1):15-25. doi: 10.1111/php.12681. Epub 2017 Jan 9. Photochem Photobiol. 2017. PMID: 27925276 Review.
Cited by
-
Kinetic Modeling of the Arabidopsis Cryptochrome Photocycle: FADH(o) Accumulation Correlates with Biological Activity.Front Plant Sci. 2016 Jun 28;7:888. doi: 10.3389/fpls.2016.00888. eCollection 2016. Front Plant Sci. 2016. PMID: 27446119 Free PMC article.
-
Human cryptochrome-1 confers light independent biological activity in transgenic Drosophila correlated with flavin radical stability.PLoS One. 2012;7(3):e31867. doi: 10.1371/journal.pone.0031867. Epub 2012 Mar 12. PLoS One. 2012. PMID: 22427812 Free PMC article.
-
Molecular Cloning and Expression Analysis of the Cryptochrome Gene CiPlant-CRY1 in Antarctic Ice Alga Chlamydomonas sp. ICE-L.Plants (Basel). 2022 Aug 26;11(17):2213. doi: 10.3390/plants11172213. Plants (Basel). 2022. PMID: 36079595 Free PMC article.
-
Magnetic sensitivity mediated by the Arabidopsis blue-light receptor cryptochrome occurs during flavin reoxidation in the dark.Planta. 2019 Feb;249(2):319-332. doi: 10.1007/s00425-018-3002-y. Epub 2018 Sep 7. Planta. 2019. PMID: 30194534
-
Plant Cryptochromes Illuminated: A Spectroscopic Perspective on the Mechanism.Front Chem. 2021 Nov 24;9:780199. doi: 10.3389/fchem.2021.780199. eCollection 2021. Front Chem. 2021. PMID: 34900940 Free PMC article. Review.
References
-
- Cashmore AR, Jarillo JA, Wu Y, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. - PubMed
-
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. - PubMed
-
- Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang R, Todo T, et al. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–13877. - PubMed
-
- Song S, Őztűrk N, Denaro TR, Arat NŐ, Kao Y, Zhu H, et al. Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J Biol Chem. 2007;282:17608–17612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

