Root-localized phytochrome chromophore synthesis is required for photoregulation of root elongation and impacts root sensitivity to jasmonic acid in Arabidopsis
- PMID: 21875894
- PMCID: PMC3252167
- DOI: 10.1104/pp.111.184689
Root-localized phytochrome chromophore synthesis is required for photoregulation of root elongation and impacts root sensitivity to jasmonic acid in Arabidopsis
Abstract
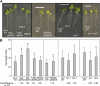
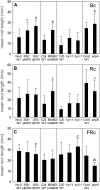
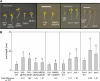
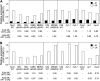
Plants exhibit organ- and tissue-specific light responses. To explore the molecular basis of spatial-specific phytochrome-regulated responses, a transgenic approach for regulating the synthesis and accumulation of the phytochrome chromophore phytochromobilin (PΦB) was employed. In prior experiments, transgenic expression of the BILIVERDIN REDUCTASE (BVR) gene was used to metabolically inactivate biliverdin IXα, a key precursor in the biosynthesis of PΦB, and thereby render cells accumulating BVR phytochrome deficient. Here, we report analyses of transgenic Arabidopsis (Arabidopsis thaliana) lines with distinct patterns of BVR accumulation dependent upon constitutive or tissue-specific, promoter-driven BVR expression that have resulted in insights on a correlation between root-localized BVR accumulation and photoregulation of root elongation. Plants with BVR accumulation in roots and a PΦB-deficient elongated hypocotyl2 (hy2-1) mutant exhibit roots that are longer than those of wild-type plants under white illumination. Additional analyses of a line with root-specific BVR accumulation generated using a GAL4-dependent bipartite enhancer-trap system confirmed that PΦB or phytochromes localized in roots directly impact light-dependent root elongation under white, blue, and red illumination. Additionally, roots of plants with constitutive plastid-localized or root-specific cytosolic BVR accumulation, as well as phytochrome chromophore-deficient hy1-1 and hy2-1 mutants, exhibit reduced sensitivity to the plant hormone jasmonic acid (JA) in JA-dependent root inhibition assays, similar to the response observed for the JA-insensitive mutants jar1 and myc2. Our analyses of lines with root-localized phytochrome deficiency or root-specific phytochrome depletion have provided novel insights into the roles of root-specific PΦB, or phytochromes themselves, in the photoregulation of root development and root sensitivity to JA.
Figures



Similar articles
-
Investigating tissue- and organ-specific phytochrome responses using FACS-assisted cell-type specific expression profiling in Arabidopsis thaliana.J Vis Exp. 2010 May 29;(39):1925. doi: 10.3791/1925. J Vis Exp. 2010. PMID: 20517200 Free PMC article.
-
Phytochrome-induced SIG2 expression contributes to photoregulation of phytochrome signalling and photomorphogenesis in Arabidopsis thaliana.J Exp Bot. 2013 Dec;64(18):5457-72. doi: 10.1093/jxb/ert308. Epub 2013 Sep 27. J Exp Bot. 2013. PMID: 24078666 Free PMC article.
-
Detection of spatial-specific phytochrome responses using targeted expression of biliverdin reductase in Arabidopsis.Plant Physiol. 2009 Jan;149(1):424-33. doi: 10.1104/pp.108.127050. Epub 2008 Oct 29. Plant Physiol. 2009. PMID: 18971430 Free PMC article.
-
Haem oxygenase: A functionally diverse enzyme of photosynthetic organisms and its role in phytochrome chromophore biosynthesis, cellular signalling and defence mechanisms.Plant Cell Environ. 2018 Mar;41(3):483-500. doi: 10.1111/pce.13116. Epub 2018 Feb 6. Plant Cell Environ. 2018. PMID: 29220548 Review.
-
Molecular interaction of jasmonate and phytochrome A signalling.J Exp Bot. 2014 Jun;65(11):2847-57. doi: 10.1093/jxb/eru230. Epub 2014 May 27. J Exp Bot. 2014. PMID: 24868039 Review.
Cited by
-
Light as stress factor to plant roots - case of root halotropism.Front Plant Sci. 2014 Dec 12;5:718. doi: 10.3389/fpls.2014.00718. eCollection 2014. Front Plant Sci. 2014. PMID: 25566292 Free PMC article.
-
Investigating tissue- and organ-specific phytochrome responses using FACS-assisted cell-type specific expression profiling in Arabidopsis thaliana.J Vis Exp. 2010 May 29;(39):1925. doi: 10.3791/1925. J Vis Exp. 2010. PMID: 20517200 Free PMC article.
-
Phytochrome A Regulates Carbon Flux in Dark Grown Tomato Seedlings.Front Plant Sci. 2019 Feb 27;10:152. doi: 10.3389/fpls.2019.00152. eCollection 2019. Front Plant Sci. 2019. PMID: 30873186 Free PMC article.
-
Mesophyll-specific phytochromes impact chlorophyll light-harvesting complexes (LHCs) and non-photochemical quenching.Plant Signal Behav. 2019;14(7):1609857. doi: 10.1080/15592324.2019.1609857. Epub 2019 Apr 30. Plant Signal Behav. 2019. PMID: 31037997 Free PMC article.
-
Underground roots monitor aboveground environment by sensing stem-piped light.Commun Integr Biol. 2016 Dec 9;9(6):e1261769. doi: 10.1080/19420889.2016.1261769. eCollection 2016. Commun Integr Biol. 2016. PMID: 28042383 Free PMC article.
References
-
- Ahmad M, Cashmore AR. (1997) The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J 11: 421–427 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Berger S, Bell E, Sadka A, Mullet JE. (1995) Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol 27: 933–942 - PubMed
-
- Bou-Torrent J, Roig-Villanova I, Martínez-García JF. (2008) Light signaling: back to space. Trends Plant Sci 13: 108–114 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

