Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/β-catenin signaling
- PMID: 21875946
- PMCID: PMC3171123
- DOI: 10.1083/jcb.201107021
Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/β-catenin signaling
Abstract
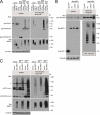
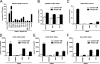
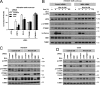
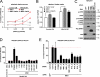
Receptor-like tyrosine kinase (RYK) functions as a transmembrane receptor for the Wnt family of secreted protein ligands. Although RYK undergoes endocytosis in response to Wnt, the mechanisms that regulate its internalization and concomitant activation of Wnt signaling are unknown. We discovered that RYK both physically and functionally interacts with the E3 ubiquitin ligase Mindbomb 1 (MIB1). Overexpression of MIB1 promotes the ubiquitination of RYK and reduces its steady-state levels at the plasma membrane. Moreover, we show that MIB1 is sufficient to activate Wnt/β-catenin (CTNNB1) signaling and that this activity depends on endogenous RYK. Conversely, in loss-of-function studies, both RYK and MIB1 are required for Wnt-3A-mediated activation of CTNNB1. Finally, we identify the Caenorhabditis elegans orthologue of MIB1 and demonstrate a genetic interaction between ceMIB and lin-18/RYK in vulva development. These findings provide insights into the mechanisms of Wnt/RYK signaling and point to novel targets for the modulation of Wnt signaling.
Figures





References
-
- Angers S., Moon R.T. 2009. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10:468–477 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

