GCC185 plays independent roles in Golgi structure maintenance and AP-1-mediated vesicle tethering
- PMID: 21875948
- PMCID: PMC3171126
- DOI: 10.1083/jcb.201104019
GCC185 plays independent roles in Golgi structure maintenance and AP-1-mediated vesicle tethering
Abstract
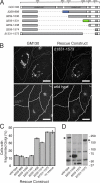
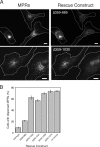
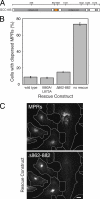
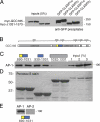
GCC185 is a long coiled-coil protein localized to the trans-Golgi network (TGN) that functions in maintaining Golgi structure and tethering mannose 6-phosphate receptor (MPR)-containing transport vesicles en route to the Golgi. We report the identification of two distinct domains of GCC185 needed either for Golgi structure maintenance or transport vesicle tethering, demonstrating the independence of these two functions. The domain needed for vesicle tethering binds to the clathrin adaptor AP-1, and cells depleted of GCC185 accumulate MPRs in transport vesicles that are AP-1 decorated. This study supports a previously proposed role of AP-1 in retrograde transport of MPRs from late endosomes to the Golgi and indicates that docking may involve the interaction of vesicle-associated AP-1 protein with the TGN-associated tethering protein GCC185.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

