Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway
- PMID: 21875955
- PMCID: PMC3182062
- DOI: 10.1084/jem.20110198
Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway
Abstract
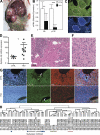
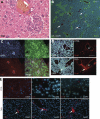
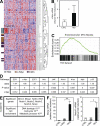
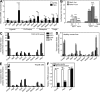
Hepatocellular carcinoma (HCC) is the third cancer killer worldwide with >600,000 deaths every year. Although the major risk factors are known, therapeutic options in patients remain limited in part because of our incomplete understanding of the cellular and molecular mechanisms influencing HCC development. Evidence indicates that the retinoblastoma (RB) pathway is functionally inactivated in most cases of HCC by genetic, epigenetic, and/or viral mechanisms. To investigate the functional relevance of this observation, we inactivated the RB pathway in the liver of adult mice by deleting the three members of the Rb (Rb1) gene family: Rb, p107, and p130. Rb family triple knockout mice develop liver tumors with histopathological features and gene expression profiles similar to human HCC. In this mouse model, cancer initiation is associated with the specific expansion of populations of liver stem/progenitor cells, indicating that the RB pathway may prevent HCC development by maintaining the quiescence of adult liver progenitor cells. In addition, we show that during tumor progression, activation of the Notch pathway via E2F transcription factors serves as a negative feedback mechanism to slow HCC growth. The level of Notch activity is also able to predict survival of HCC patients, suggesting novel means to diagnose and treat HCC.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

