Convergent evolution of two mammalian neuronal enhancers by sequential exaptation of unrelated retroposons
- PMID: 21876128
- PMCID: PMC3174587
- DOI: 10.1073/pnas.1104997108
Convergent evolution of two mammalian neuronal enhancers by sequential exaptation of unrelated retroposons
Erratum in
- Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20850
Abstract
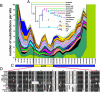
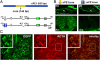
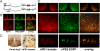
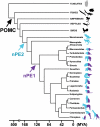
The proopiomelanocortin gene (POMC) is expressed in a group of neurons present in the arcuate nucleus of the hypothalamus. Neuron-specific POMC expression in mammals is conveyed by two distal enhancers, named nPE1 and nPE2. Previous transgenic mouse studies showed that nPE1 and nPE2 independently drive reporter gene expression to POMC neurons. Here, we investigated the evolutionary mechanisms that shaped not one but two neuron-specific POMC enhancers and tested whether nPE1 and nPE2 drive identical or complementary spatiotemporal expression patterns. Sequence comparison among representative genomes of most vertebrate classes and mammalian orders showed that nPE1 is a placental novelty. Using in silico paleogenomics we found that nPE1 originated from the exaptation of a mammalian-apparent LTR retrotransposon sometime between the metatherian/eutherian split (147 Mya) and the placental mammal radiation (≈ 90 Mya). Thus, the evolutionary origin of nPE1 differs, in kind and time, from that previously demonstrated for nPE2, which was exapted from a CORE-short interspersed nucleotide element (SINE) retroposon before the origin of prototherians, 166 Mya. Transgenic mice expressing the fluorescent markers tomato and EGFP driven by nPE1 or nPE2, respectively, demonstrated coexpression of both reporter genes along the entire arcuate nucleus. The onset of reporter gene expression guided by nPE1 and nPE2 was also identical and coincidental with the onset of Pomc expression in the presumptive mouse diencephalon. Thus, the independent exaptation of two unrelated retroposons into functional analogs regulating neuronal POMC expression constitutes an authentic example of convergent molecular evolution of cell-specific enhancers.
Conflict of interest statement
Conflict of interest statement: M.J.L., F.J.S.d.S., and M.R. have intellectual property and patent interests in the POMC neuronal-specific enhancers and have received income from the licensing of this intellectual property and related research material to financially interested companies.
Figures

Similar articles
-
Partially redundant enhancers cooperatively maintain Mammalian pomc expression above a critical functional threshold.PLoS Genet. 2015 Feb 11;11(2):e1004935. doi: 10.1371/journal.pgen.1004935. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25671638 Free PMC article.
-
Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene.PLoS Genet. 2007 Oct;3(10):1813-26. doi: 10.1371/journal.pgen.0030166. PLoS Genet. 2007. PMID: 17922573 Free PMC article.
-
A mammalian tripartite enhancer cluster controls hypothalamic Pomc expression, food intake, and body weight.Proc Natl Acad Sci U S A. 2024 Apr 30;121(18):e2322692121. doi: 10.1073/pnas.2322692121. Epub 2024 Apr 23. Proc Natl Acad Sci U S A. 2024. PMID: 38652744 Free PMC article.
-
Molecular and functional genetics of the proopiomelanocortin gene, food intake regulation and obesity.FEBS Lett. 2017 Sep;591(17):2593-2606. doi: 10.1002/1873-3468.12776. Epub 2017 Aug 20. FEBS Lett. 2017. PMID: 28771698 Free PMC article. Review.
-
The Phylogeny of Placental Evolution Through Dynamic Integrations of Retrotransposons.Prog Mol Biol Transl Sci. 2017;145:89-109. doi: 10.1016/bs.pmbts.2016.12.004. Epub 2017 Jan 16. Prog Mol Biol Transl Sci. 2017. PMID: 28110755 Review.
Cited by
-
Introns: the "dark matter" of the eukaryotic genome.Front Genet. 2023 May 16;14:1150212. doi: 10.3389/fgene.2023.1150212. eCollection 2023. Front Genet. 2023. PMID: 37260773 Free PMC article. Review.
-
Alu elements and human common diseases like obesity.Mob Genet Elements. 2012 Jul 1;2(4):197-201. doi: 10.4161/mge.21470. Mob Genet Elements. 2012. PMID: 23087845 Free PMC article.
-
Partially redundant enhancers cooperatively maintain Mammalian pomc expression above a critical functional threshold.PLoS Genet. 2015 Feb 11;11(2):e1004935. doi: 10.1371/journal.pgen.1004935. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25671638 Free PMC article.
-
Maternal overnutrition programs epigenetic changes in the regulatory regions of hypothalamic Pomc in the offspring of rats.Int J Obes (Lond). 2018 Aug;42(8):1431-1444. doi: 10.1038/s41366-018-0094-1. Epub 2018 May 17. Int J Obes (Lond). 2018. PMID: 29777232 Free PMC article.
-
Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates.Chromosome Res. 2015 Sep;23(3):505-31. doi: 10.1007/s10577-015-9493-5. Chromosome Res. 2015. PMID: 26395902 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

