Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions
- PMID: 21876132
- PMCID: PMC3174574
- DOI: 10.1073/pnas.1018949108
Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions
Abstract
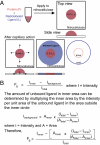
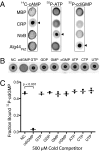
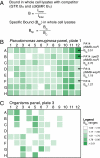
Interactions of proteins with low-molecular-weight ligands, such as metabolites, cofactors, and allosteric regulators, are important determinants of metabolism, gene regulation, and cellular homeostasis. Pharmaceuticals often target these interactions to interfere with regulatory pathways. We have developed a rapid, precise, and high-throughput method for quantitatively measuring protein-ligand interactions without the need to purify the protein when performed in cells with low background activity. This method, differential radial capillary action of ligand assay (DRaCALA), is based on the ability of dry nitrocellulose to separate the free ligand from bound protein-ligand complexes. Nitrocellulose sequesters proteins and bound ligand at the site of application, whereas free ligand is mobilized by bulk movement of the solvent through capillary action. We show here that DRaCALA allows detection of specific interactions between three nucleotides and their cognate binding proteins. DRaCALA allows quantitative measurement of the dissociation constant and the dissociation rate. Furthermore, DRaCALA can detect the expression of a cyclic-di-GMP (cdiGMP)-binding protein in whole-cell lysates of Escherichia coli, demonstrating the power of the method to bypass the prerequisite for protein purification. We have used DRaCALA to investigate cdiGMP signaling in 54 bacterial species from 37 genera and 7 eukaryotic species. These studies revealed the presence of potential cdiGMP-binding proteins in 21 species of bacteria, including 4 unsequenced species. The ease of obtaining metabolite-protein interaction data using the DRaCALA assay will facilitate rapid identification of protein-metabolite and protein-pharmaceutical interactions in a systematic and comprehensive approach.
Conflict of interest statement
Conflict of interest statement: A provisional patent has been filed by the University of Maryland.
Figures


Similar articles
-
High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases.ACS Chem Biol. 2014 Jan 17;9(1):183-92. doi: 10.1021/cb400485k. Epub 2013 Nov 4. ACS Chem Biol. 2014. PMID: 24134695 Free PMC article.
-
Differential Radial Capillary Action of Ligand Assay (DRaCALA) for High-Throughput Detection of Protein-Metabolite Interactions in Bacteria.Methods Mol Biol. 2017;1535:25-41. doi: 10.1007/978-1-4939-6673-8_3. Methods Mol Biol. 2017. PMID: 27914071
-
Differential Radial Capillary Action of Ligand Assay (DRaCALA).Curr Protoc Mol Biol. 2019 Apr;126(1):e84. doi: 10.1002/cpmb.84. Epub 2018 Dec 3. Curr Protoc Mol Biol. 2019. PMID: 30508276 Free PMC article.
-
[Identification of cyclic di-GMP protein receptors: high-throughput screening strategies and experimental verification].Sheng Wu Gong Cheng Xue Bao. 2017 Sep 25;33(9):1376-1389. doi: 10.13345/j.cjb.170175. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 28956389 Review. Chinese.
-
[Mode of action of cyclic amp in prokaryotes and eukaryotes, CAP and cAMP-dependent protein kinases].Biochimie. 1985 Jun;67(6):563-82. doi: 10.1016/s0300-9084(85)80196-6. Biochimie. 1985. PMID: 2413906 Review. French.
Cited by
-
Detection of cyclic diguanylate G-octaplex assembly and interaction with proteins.PLoS One. 2013;8(1):e53689. doi: 10.1371/journal.pone.0053689. Epub 2013 Jan 7. PLoS One. 2013. PMID: 23308275 Free PMC article.
-
Gut colonization by Bacteroides requires translation by an EF-G paralog lacking GTPase activity.EMBO J. 2023 Jan 16;42(2):e112372. doi: 10.15252/embj.2022112372. Epub 2022 Dec 6. EMBO J. 2023. PMID: 36472247 Free PMC article.
-
Core Transmembrane Domain 6 Plays a Pivotal Role in the Transport Cycle of the Sodium/Proline Symporter PutP.J Biol Chem. 2016 Dec 9;291(50):26208-26215. doi: 10.1074/jbc.M116.753103. Epub 2016 Oct 28. J Biol Chem. 2016. PMID: 27793991 Free PMC article.
-
Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.Nat Commun. 2022 Oct 3;13(1):5834. doi: 10.1038/s41467-022-33537-w. Nat Commun. 2022. PMID: 36192422 Free PMC article.
-
A Subset of Exoribonucleases Serve as Degradative Enzymes for pGpG in c-di-GMP Signaling.J Bacteriol. 2018 Nov 26;200(24):e00300-18. doi: 10.1128/JB.00300-18. Print 2018 Dec 15. J Bacteriol. 2018. PMID: 30249708 Free PMC article.
References
-
- Lewin B. Cells. Sudbury, MA: Jones and Bartlett Publishers; 2007. p. xix.
-
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. - PubMed
-
- Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

