Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding
- PMID: 21876142
- PMCID: PMC3174611
- DOI: 10.1073/pnas.1106341108
Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding
Abstract
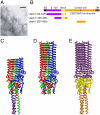
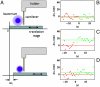
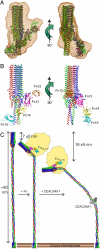
Bacterial cell surfaces are commonly decorated with a layer formed from multiple copies of adhesin proteins whose binding interactions initiate colonization and infection processes. In this study, we investigate the physical deformability of the UspA1 adhesin protein from Moraxella catarrhalis, a causative agent of middle-ear infections in humans. UspA1 binds a range of extracellular proteins including fibronectin, and the epithelial cellular receptor carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1). Electron microscopy indicates that unliganded UspA1 is densely packed at, and extends about 800 Å from, the Moraxella surface. Using a modified atomic force microscope, we show that the adhesive properties and thickness of the UspA1 layer at the cell surface varies on addition of either fibronectin or CEACAM1. This in situ analysis is then correlated with the molecular structure of UspA1. To provide an overall model for UspA1, we have determined crystal structures for two N-terminal fragments which are then combined with a previous structure of the CEACAM1-binding site. We show that the UspA1-fibronectin complex is formed between UspA1 head region and the 13th type-III domain of fibronectin and, using X-ray scattering, that the complex involves an angular association between these two proteins. In combination with a previous study, which showed that the CEACAM1-UspA1 complex is distinctively bent in solution, we correlate these observations on isolated fragments of UspA1 with its in situ response on the cell surface. This study therefore provides a rare direct demonstration of protein conformational change at the cell surface.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Karalus R, Campagnari A. Moraxella catarrhalis: A review of an important human mucosal pathogen. Microbes Infect. 2000;2:547–559. - PubMed
-
- Hultgren SJ, et al. Pilus and nonpilus bacterial adhesins: Assembly and function in cell recognition. Cell. 1993;73:887–901. - PubMed
-
- Wells TJ, Tree JJ, Ulett GC, Schembri MA. Autotransporter proteins: Novel targets at the bacterial cell surface. FEMS Microbiol Lett. 2007;274:163–172. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

