Functional characterization of bacterial sRNAs using a network biology approach
- PMID: 21876160
- PMCID: PMC3174649
- DOI: 10.1073/pnas.1104318108
Functional characterization of bacterial sRNAs using a network biology approach
Abstract
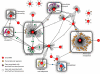
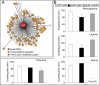
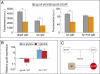
Small RNAs (sRNAs) are important components of posttranscriptional regulation. These molecules are prevalent in bacterial and eukaryotic organisms, and involved in a variety of responses to environmental stresses. The functional characterization of sRNAs is challenging and requires highly focused and extensive experimental procedures. Here, using a network biology approach and a compendium of gene expression profiles, we predict functional roles and regulatory interactions for sRNAs in Escherichia coli. We experimentally validate predictions for three sRNAs in our inferred network: IsrA, GlmZ, and GcvB. Specifically, we validate a predicted role for IsrA and GlmZ in the SOS response, and we expand on current knowledge of the GcvB sRNA, demonstrating its broad role in the regulation of amino acid metabolism and transport. We also show, using the inferred network coupled with experiments, that GcvB and Lrp, a transcription factor, repress each other in a mutually inhibitory network. This work shows that a network-based approach can be used to identify the cellular function of sRNAs and characterize the relationship between sRNAs and transcription factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

