Observation of spatial propagation of amyloid assembly from single nuclei
- PMID: 21876182
- PMCID: PMC3169119
- DOI: 10.1073/pnas.1105555108
Observation of spatial propagation of amyloid assembly from single nuclei
Abstract
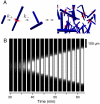
The crucial early stages of amyloid growth, in which normally soluble proteins are converted into fibrillar nanostructures, are challenging to study using conventional techniques yet are critical to the protein aggregation phenomena implicated in many common pathologies. As with all nucleation and growth phenomena, it is difficult to track individual nuclei in traditional macroscopic experiments, which probe the overall temporal evolution of the sample, but do not yield detailed information on the primary nucleation step as they mix independent stochastic events into an ensemble measurement. To overcome this limitation, we have developed microdroplet assays enabling us to detect single primary nucleation events and to monitor their subsequent spatial as well as temporal evolution, both of which we find to be determined by secondary nucleation phenomena. By deforming the droplets to high aspect ratio, we visualize in real-time propagating waves of protein assembly emanating from discrete primary nucleation sites. We show that, in contrast to classical gelation phenomena, the primary nucleation step is characterized by a striking dependence on system size, and the filamentous protein self-assembly process involves a highly nonuniform spatial distribution of aggregates. These findings deviate markedly from the current picture of amyloid growth and uncover a general driving force, originating from confinement, which, together with biological quality control mechanisms, helps proteins remain soluble and therefore functional in nature.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Self-assembly of Mutant Huntingtin Exon-1 Fragments into Large Complex Fibrillar Structures Involves Nucleated Branching.J Mol Biol. 2018 Jun 8;430(12):1725-1744. doi: 10.1016/j.jmb.2018.03.017. Epub 2018 Mar 28. J Mol Biol. 2018. PMID: 29601786
-
On the lag phase in amyloid fibril formation.Phys Chem Chem Phys. 2015 Mar 28;17(12):7606-18. doi: 10.1039/c4cp05563b. Phys Chem Chem Phys. 2015. PMID: 25719972 Free PMC article. Review.
-
Connecting macroscopic observables and microscopic assembly events in amyloid formation using coarse grained simulations.PLoS Comput Biol. 2012;8(10):e1002692. doi: 10.1371/journal.pcbi.1002692. Epub 2012 Oct 11. PLoS Comput Biol. 2012. PMID: 23071427 Free PMC article.
-
Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism.Proc Natl Acad Sci U S A. 2013 Jun 11;110(24):9758-63. doi: 10.1073/pnas.1218402110. Epub 2013 May 23. Proc Natl Acad Sci U S A. 2013. PMID: 23703910 Free PMC article.
-
The Nucleation of Protein Aggregates - From Crystals to Amyloid Fibrils.Int Rev Cell Mol Biol. 2017;329:187-226. doi: 10.1016/bs.ircmb.2016.08.014. Epub 2016 Oct 22. Int Rev Cell Mol Biol. 2017. PMID: 28109328 Review.
Cited by
-
Toward the quantification of α-synuclein aggregates with digital seed amplification assays.Proc Natl Acad Sci U S A. 2024 Jan 16;121(3):e2312031121. doi: 10.1073/pnas.2312031121. Epub 2024 Jan 9. Proc Natl Acad Sci U S A. 2024. PMID: 38194461 Free PMC article.
-
Distinct thermodynamic signatures of oligomer generation in the aggregation of the amyloid-β peptide.Nat Chem. 2018 May;10(5):523-531. doi: 10.1038/s41557-018-0023-x. Epub 2018 Mar 26. Nat Chem. 2018. PMID: 29581486 Free PMC article.
-
Protein Misfolding in Pregnancy: Current Insights, Potential Mechanisms, and Implications for the Pathogenesis of Preeclampsia.Molecules. 2024 Jan 26;29(3):610. doi: 10.3390/molecules29030610. Molecules. 2024. PMID: 38338354 Free PMC article. Review.
-
Cyclic undecapeptide Cyclosporin A mediated inhibition of amyloid synthesis: Implications in alleviation of amyloid induced neurotoxicity.Sci Rep. 2018 Nov 23;8(1):17283. doi: 10.1038/s41598-018-35645-4. Sci Rep. 2018. PMID: 30470780 Free PMC article.
-
Biophysical Aspects of Alzheimer's Disease: Implications for Pharmaceutical Sciences : Theme: Drug Discovery, Development and Delivery in Alzheimer's Disease Guest Editor: Davide Brambilla.Pharm Res. 2017 Dec;34(12):2628-2636. doi: 10.1007/s11095-017-2266-4. Epub 2017 Sep 28. Pharm Res. 2017. PMID: 28963701 Review.
References
-
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. - PubMed
-
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid-from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. - PubMed
-
- Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. - PubMed
-
- Sacchettini JC, Kelly JW. Therapeutic strategies for human amyloid diseases. Nat Rev Drug Discov. 2002;1:267–275. - PubMed
-
- Westermark P. Amyloid Proteins. Vol. 3. Weinheim: Wiley; 2005. (Amyloidosis and Amyloid Proteins: Brief History and Definitions).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

