Reiterative Recombination for the in vivo assembly of libraries of multigene pathways
- PMID: 21876185
- PMCID: PMC3174608
- DOI: 10.1073/pnas.1100507108
Reiterative Recombination for the in vivo assembly of libraries of multigene pathways
Abstract
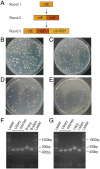
The increasing sophistication of synthetic biology is creating a demand for robust, broadly accessible methodology for constructing multigene pathways inside of the cell. Due to the difficulty of rationally designing pathways that function as desired in vivo, there is a further need to assemble libraries of pathways in parallel, in order to facilitate the combinatorial optimization of performance. While some in vitro DNA assembly methods can theoretically make libraries of pathways, these techniques are resource intensive and inherently require additional techniques to move the DNA back into cells. All previously reported in vivo assembly techniques have been low yielding, generating only tens to hundreds of constructs at a time. Here, we develop "Reiterative Recombination," a robust method for building multigene pathways directly in the yeast chromosome. Due to its use of endonuclease-induced homologous recombination in conjunction with recyclable markers, Reiterative Recombination provides a highly efficient, technically simple strategy for sequentially assembling an indefinite number of DNA constructs at a defined locus. In this work, we describe the design and construction of the first Reiterative Recombination system in Saccharomyces cerevisiae, and we show that it can be used to assemble multigene constructs. We further demonstrate that Reiterative Recombination can construct large mock libraries of at least 10(4) biosynthetic pathways. We anticipate that our system's simplicity and high efficiency will make it a broadly accessible technology for pathway construction and render it a valuable tool for optimizing pathways in vivo.
Conflict of interest statement
Conflict of interest statement: L.M.W. and V.W.C. are inventors on a patent filed regarding this method.
Figures


Similar articles
-
Gene assembly and combinatorial libraries in S. cerevisiae via reiterative recombination.Methods Mol Biol. 2013;978:187-203. doi: 10.1007/978-1-62703-293-3_14. Methods Mol Biol. 2013. PMID: 23423898
-
Automated Construction of a Yeast-Based Multigene Library via Homologous Recombination in a Biofoundry Workflow.ACS Synth Biol. 2025 May 16;14(5):1549-1556. doi: 10.1021/acssynbio.4c00812. Epub 2025 May 7. ACS Synth Biol. 2025. PMID: 40331904
-
Construction and engineering of large biochemical pathways via DNA assembler.Methods Mol Biol. 2013;1073:85-106. doi: 10.1007/978-1-62703-625-2_9. Methods Mol Biol. 2013. PMID: 23996442 Free PMC article.
-
Recombination in yeast and the recombinant DNA technology.Genome. 1989;31(2):536-40. doi: 10.1139/g89-102. Genome. 1989. PMID: 2698829 Review.
-
Design-based re-engineering of biosynthetic gene clusters: plug-and-play in practice.Curr Opin Biotechnol. 2013 Dec;24(6):1144-50. doi: 10.1016/j.copbio.2013.03.006. Epub 2013 Mar 27. Curr Opin Biotechnol. 2013. PMID: 23540422 Review.
Cited by
-
A Heritable Recombination system for synthetic Darwinian evolution in yeast.ACS Synth Biol. 2012 Dec 21;1(12):602-9. doi: 10.1021/sb3000904. ACS Synth Biol. 2012. PMID: 23412545 Free PMC article.
-
Total Heterologous Biosynthesis of Fungal Natural Products in Aspergillus nidulans.J Nat Prod. 2022 Oct 28;85(10):2484-2518. doi: 10.1021/acs.jnatprod.2c00487. Epub 2022 Sep 29. J Nat Prod. 2022. PMID: 36173392 Free PMC article. Review.
-
Utilization of a calmodulin lysine methyltransferase co-expression system for the generation of a combinatorial library of post-translationally modified proteins.Protein Expr Purif. 2012 Dec;86(2):83-8. doi: 10.1016/j.pep.2012.09.012. Epub 2012 Oct 2. Protein Expr Purif. 2012. PMID: 23036357 Free PMC article.
-
Draft Genome Sequence of Saccharomyces cerevisiae LW2591Y, a Laboratory Strain for In Vivo Multigene Assemblies.Microbiol Resour Announc. 2021 Mar 4;10(9):e01418-20. doi: 10.1128/MRA.01418-20. Microbiol Resour Announc. 2021. PMID: 33664139 Free PMC article.
-
Recent advances in DNA assembly technologies.FEMS Yeast Res. 2015 Feb;15(1):1-9. doi: 10.1111/1567-1364.12171. Epub 2015 Jan 14. FEMS Yeast Res. 2015. PMID: 24903193 Free PMC article. Review.
References
-
- Baker D, et al. Engineering life: building a fab for biology. Sci Am. 2006;294:44–51. - PubMed
-
- Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. - PubMed
-
- Haseltine EL, Arnold FH. Synthetic gene circuits: design with directed evolution. Annu Rev Biophys Biomol Struct. 2007;36:1–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

