Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1)
- PMID: 21876670
- PMCID: PMC3158049
- DOI: 10.1371/journal.ppat.1002184
Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1)
Abstract
The essential transactivator function of the HIV Tat protein is regulated by multiple posttranslational modifications. Although individual modifications are well characterized, their crosstalk and dynamics of occurrence during the HIV transcription cycle remain unclear.We examine interactions between two critical modifications within the RNA-binding domain of Tat: monomethylation of lysine 51 (K51) mediated by Set7/9/KMT7, an early event in the Tat transactivation cycle that strengthens the interaction of Tat with TAR RNA, and acetylation of lysine 50 (K50) mediated by p300/KAT3B, a later process that dissociates the complex formed by Tat, TAR RNA and the cyclin T1 subunit of the positive transcription elongation factor b (P-TEFb). We find K51 monomethylation inhibited in synthetic Tat peptides carrying an acetyl group at K50 while acetylation can occur in methylated peptides, albeit at a reduced rate. To examine whether Tat is subject to sequential monomethylation and acetylation in cells, we performed mass spectrometry on immunoprecipitated Tat proteins and generated new modification-specific Tat antibodies against monomethylated/acetylated Tat. No bimodified Tat protein was detected in cells pointing to a demethylation step during the Tat transactivation cycle. We identify lysine-specific demethylase 1 (LSD1/KDM1) as a Tat K51-specific demethylase, which is required for the activation of HIV transcription in latently infected T cells. LSD1/KDM1 and its cofactor CoREST associates with the HIV promoter in vivo and activate Tat transcriptional activity in a K51-dependent manner. In addition, small hairpin RNAs directed against LSD1/KDM1 or inhibition of its activity with the monoamine oxidase inhibitor phenelzine suppresses the activation of HIV transcription in latently infected T cells.Our data support the model that a LSD1/KDM1/CoREST complex, normally known as a transcriptional suppressor, acts as a novel activator of HIV transcription through demethylation of K51 in Tat. Small molecule inhibitors of LSD1/KDM1 show therapeutic promise by enforcing HIV latency in infected T cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

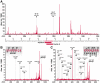



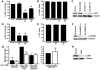


References
-
- Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5:95–106. - PubMed
-
- Noe A, Plum J, Verhofstede C. The latent HIV-1 reservoir in patients undergoing HAART: an archive of pre-HAART drug resistance. J Antimicrob Chemother. 2005;55:410–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

