Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea
- PMID: 21876677
- PMCID: PMC3158057
- DOI: 10.1371/journal.pgen.1002230
Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea
Abstract
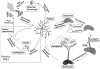
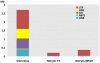
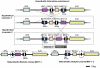
Sclerotinia sclerotiorum and Botrytis cinerea are closely related necrotrophic plant pathogenic fungi notable for their wide host ranges and environmental persistence. These attributes have made these species models for understanding the complexity of necrotrophic, broad host-range pathogenicity. Despite their similarities, the two species differ in mating behaviour and the ability to produce asexual spores. We have sequenced the genomes of one strain of S. sclerotiorum and two strains of B. cinerea. The comparative analysis of these genomes relative to one another and to other sequenced fungal genomes is provided here. Their 38-39 Mb genomes include 11,860-14,270 predicted genes, which share 83% amino acid identity on average between the two species. We have mapped the S. sclerotiorum assembly to 16 chromosomes and found large-scale co-linearity with the B. cinerea genomes. Seven percent of the S. sclerotiorum genome comprises transposable elements compared to <1% of B. cinerea. The arsenal of genes associated with necrotrophic processes is similar between the species, including genes involved in plant cell wall degradation and oxalic acid production. Analysis of secondary metabolism gene clusters revealed an expansion in number and diversity of B. cinerea-specific secondary metabolites relative to S. sclerotiorum. The potential diversity in secondary metabolism might be involved in adaptation to specific ecological niches. Comparative genome analysis revealed the basis of differing sexual mating compatibility systems between S. sclerotiorum and B. cinerea. The organization of the mating-type loci differs, and their structures provide evidence for the evolution of heterothallism from homothallism. These data shed light on the evolutionary and mechanistic bases of the genetically complex traits of necrotrophic pathogenicity and sexual mating. This resource should facilitate the functional studies designed to better understand what makes these fungi such successful and persistent pathogens of agronomic crops.
Conflict of interest statement
I have read the journal's policy and have the following conflicts: author Chinnappa Kodira currently works at 454 Life Sciences, Roche. All of the work reported in this manuscript was completed when he was in residence at the Broad Institute. None of the other authors have declared any competing interests.
Figures

References
-
- Bolton MD, Thomma BP, Nelson BD. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol. 2006;7:1–16. - PubMed
-
- Williamson B, Tudzynski B, Tudzynski P, van Kan JAL. Botrytis cinerea: The cause of grey mould disease. Mol Plant Pathol. 2007;8:561–580. - PubMed
-
- Oliver RP, Solomon PS. New developments in pathogenicity and virulence of necrotrophs. Curr Opin Plant Biol. 2010;13:415–419. - PubMed
-
- Kim KS, Min JY, Dickman MB. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol Plant Microbe Interact. 2008;21:605–612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

