Protein-binding microarray analysis of tumor suppressor AP2α target gene specificity
- PMID: 21876733
- PMCID: PMC3158074
- DOI: 10.1371/journal.pone.0022895
Protein-binding microarray analysis of tumor suppressor AP2α target gene specificity
Abstract
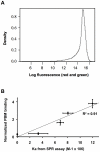
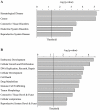
Cheap and massively parallel methods to assess the DNA-binding specificity of transcription factors are actively sought, given their prominent regulatory role in cellular processes and diseases. Here we evaluated the use of protein-binding microarrays (PBM) to probe the association of the tumor suppressor AP2α with 6000 human genomic DNA regulatory sequences. We show that the PBM provides accurate relative binding affinities when compared to quantitative surface plasmon resonance assays. A PBM-based study of human healthy and breast tumor tissue extracts allowed the identification of previously unknown AP2α target genes and it revealed genes whose direct or indirect interactions with AP2α are affected in the diseased tissues. AP2α binding and regulation was confirmed experimentally in human carcinoma cells for novel target genes involved in tumor progression and resistance to chemotherapeutics, providing a molecular interpretation of AP2α role in cancer chemoresistance. Overall, we conclude that this approach provides quantitative and accurate assays of the specificity and activity of tumor suppressor and oncogenic proteins in clinical samples, interfacing genomic and proteomic assays.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

