CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing
- PMID: 21878105
- PMCID: PMC3228541
- DOI: 10.1186/1471-2105-12-356
CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing
Abstract
Background: Next-generation sequencing technologies have decentralized sequence acquisition, increasing the demand for new bioinformatics tools that are easy to use, portable across multiple platforms, and scalable for high-throughput applications. Cloud computing platforms provide on-demand access to computing infrastructure over the Internet and can be used in combination with custom built virtual machines to distribute pre-packaged with pre-configured software.
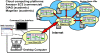
Results: We describe the Cloud Virtual Resource, CloVR, a new desktop application for push-button automated sequence analysis that can utilize cloud computing resources. CloVR is implemented as a single portable virtual machine (VM) that provides several automated analysis pipelines for microbial genomics, including 16S, whole genome and metagenome sequence analysis. The CloVR VM runs on a personal computer, utilizes local computer resources and requires minimal installation, addressing key challenges in deploying bioinformatics workflows. In addition CloVR supports use of remote cloud computing resources to improve performance for large-scale sequence processing. In a case study, we demonstrate the use of CloVR to automatically process next-generation sequencing data on multiple cloud computing platforms.
Conclusion: The CloVR VM and associated architecture lowers the barrier of entry for utilizing complex analysis protocols on both local single- and multi-core computers and cloud systems for high throughput data processing.
Figures






References
-
- Next Generation Genomics: World Map of High-throughput Sequencers. http://pathogenomics.bham.ac.uk/hts/ http://pathogenomics.bham.ac.uk/hts/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

