Cellular reprogramming by the conjoint action of ERα, FOXA1, and GATA3 to a ligand-inducible growth state
- PMID: 21878914
- PMCID: PMC3202798
- DOI: 10.1038/msb.2011.59
Cellular reprogramming by the conjoint action of ERα, FOXA1, and GATA3 to a ligand-inducible growth state
Abstract
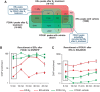
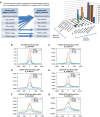
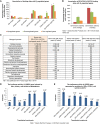
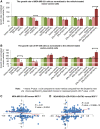
Despite the role of the estrogen receptor α (ERα) pathway as a key growth driver for breast cells, the phenotypic consequence of exogenous introduction of ERα into ERα-negative cells paradoxically has been growth inhibition. We mapped the binding profiles of ERα and its interacting transcription factors (TFs), FOXA1 and GATA3 in MCF-7 breast carcinoma cells, and observed that these three TFs form a functional enhanceosome that regulates the genes driving core ERα function and cooperatively modulate the transcriptional networks previously ascribed to ERα alone. We demonstrate that these enhanceosome occupied sites are associated with optimal enhancer characteristics with highest p300 co-activator recruitment, RNA Pol II occupancy, and chromatin opening. Most importantly, we show that the transfection of all three TFs was necessary to reprogramme the ERα-negative MDA-MB-231 and BT-549 cells to restore the estrogen-responsive growth resembling estrogen-treated ERα-positive MCF-7 cells. Cumulatively, these results suggest that all the enhanceosome components comprising ERα, FOXA1, and GATA3 are necessary for the full repertoire of cancer-associated effects of the ERα.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE (2007) Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209 - PubMed
-
- Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG, Nakshatri H (2007) FOXA1 expression in breast cancer—correlation with luminal subtype A and survival. Clin Cancer Res 13: 4415–4421 - PubMed
-
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

