Ran-dependent nuclear export mediators: a structural perspective
- PMID: 21878989
- PMCID: PMC3181476
- DOI: 10.1038/emboj.2011.287
Ran-dependent nuclear export mediators: a structural perspective
Abstract
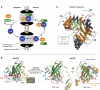
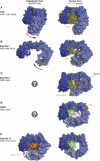
Nuclear export is an essential eukaryotic activity. It proceeds through nuclear pore complexes (NPCs) and is mediated by soluble receptors that shuttle between nucleus and cytoplasm. RanGTPase-dependent export mediators (exportins) constitute the largest class of these carriers and are functionally highly versatile. All of these exportins load their substrates in response to RanGTP binding in the nucleus and traverse NPCs as ternary RanGTP-exportin-cargo complexes to the cytoplasm, where GTP hydrolysis leads to export complex disassembly. The different exportins vary greatly in their substrate range. Recent structural studies of both protein- and RNA-specific exporters have illuminated how exportins bind their cargoes, how Ran triggers cargo loading and how export complexes are disassembled in the cytoplasm. Here, we review the current state of knowledge and highlight emerging principles as well as prevailing questions.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2007) Molecular Biology of the Cell, 5th ed. Garland Science, ISBN 0815341059
-
- Andrade MA, Bork P (1995) HEAT repeats in the Huntington's disease protein. Nat Genet 11: 115–116 - PubMed
-
- Arts GJ, Fornerod M, Mattaj IW (1998a) Identification of a nuclear export receptor for tRNA. Curr Biol 8: 305–314 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

