G-patch domain and KOW motifs-containing protein, GPKOW; a nuclear RNA-binding protein regulated by protein kinase A
- PMID: 21880142
- PMCID: PMC3179746
- DOI: 10.1186/1750-2187-6-10
G-patch domain and KOW motifs-containing protein, GPKOW; a nuclear RNA-binding protein regulated by protein kinase A
Abstract
Background: Post-transcriptional processing of pre-mRNA takes place in several steps and requires involvement of a number of RNA-binding proteins. How pre-mRNA processing is regulated is in large enigmatic. The catalytic (C) subunit of protein kinase A (PKA) is a serine/threonine kinase, which regulates numerous cellular processes including pre-mRNA splicing. Despite that a significant fraction of the C subunit is found in splicing factor compartments in the nucleus, there are no indications of a direct interaction between RNA and PKA. Based on this we speculate if the specificity of the C subunit in regulating pre-mRNA splicing may be mediated indirectly through other nuclear proteins.
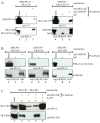
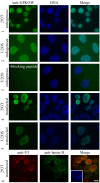
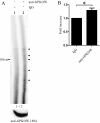
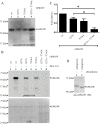
Results: Using yeast two-hybrid screening with the PKA C subunit Cbeta2 as bait, we identified the G-patch domain and KOW motifs-containing protein (GPKOW), also known as the T54 protein or MOS2 homolog, as an interaction partner for Cbeta2. We demonstrate that GPKOW, which contains one G-patch domain and two KOW motifs, is a nuclear RNA-binding protein conserved between species. GPKOW contains two sites that are phosphorylated by PKA in vitro. By RNA immunoprecipitation and site directed mutagenesis of the PKA phosphorylation sites we revealed that GPKOW binds RNA in vivo in a PKA sensitive fashion.
Conclusion: GPKOW is a RNA-binding protein that binds RNA in a PKA regulated fashion. Together with our previous results demonstrating that PKA regulates pre-mRNA splicing, our results suggest that PKA phosphorylation is involved in regulating RNA processing at several steps.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

