Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803
- PMID: 21880717
- PMCID: PMC3196108
- DOI: 10.1074/jbc.M111.263780
Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803
Erratum in
- J Biol Chem. 2012 Mar 9;287(11):8660
Abstract
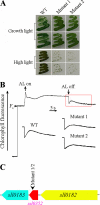
Cyanobacterial NADPH:plastoquinone oxidoreductase, or type I NAD(P)H dehydrogenase, or the NDH-1 complex is involved in plastoquinone reduction and cyclic electron transfer (CET) around photosystem I. CET, in turn, produces extra ATP for cell metabolism particularly under stressful conditions. Despite significant achievements in the study of cyanobacterial NDH-1 complexes during the past few years, the entire subunit composition still remains elusive. To identify missing subunits, we screened a transposon-tagged library of Synechocystis 6803 cells grown under high light. Two NDH-1-mediated CET (NDH-CET)-defective mutants were tagged in the same ssl0352 gene encoding a short unknown protein. To clarify the function of Ssl0352, the ssl0352 deletion mutant and another mutant with Ssl0352 fused to yellow fluorescent protein (YFP) and the His(6) tag were constructed. Immunoblotting, mass spectrometry, and confocal microscopy analyses revealed that the Ssl0352 protein resides in the thylakoid membrane and associates with the NDH-1L and NDH-1M complexes. We conclude that Ssl0352 is a novel subunit of cyanobacterial NDH-1 complexes and designate it NdhS. Deletion of the ssl0352 gene considerably impaired the NDH-CET activity and also retarded cell growth under high light conditions, indicating that NdhS is essential for efficient operation of NDH-CET. However, the assembly of the NDH-1L and NDH-1M complexes and their content in the cells were not affected in the mutant. NdhS contains a Src homology 3-like domain and might be involved in interaction of the NDH-1 complex with an electron donor.
Figures







References
-
- Mi H., Endo T., Schreiber U., Ogawa T., Asada K. (1992) Plant Cell Physiol. 33, 1233–1237
-
- Ohkawa H., Pakrasi H. B., Ogawa T. (2000) J. Biol. Chem. 275, 31630–31634 - PubMed
-
- Friedrich T., Steinmüller K., Weiss H. (1995) FEBS Lett. 367, 107–111 - PubMed
-
- Friedrich T., Scheide D. (2000) FEBS Lett. 479, 1–5 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

